Abstract
Understanding where proteins are localized in a bacterial cell is essential for understanding their function and regulation. This is particularly important for proteins that are involved in cell division, which localize at the division septum and assemble into highly regulated complexes. Current knowledge of these complexes has been greatly facilitated by super-resolution imaging using fluorescent protein fusions. Herein, we demonstrate with FtsZ that single-molecule PALM images can be obtained in-vivo using a genetically fused nanotag (ALFA), and a corresponding nanobody fused to mEos3.2. The methodology presented is applicable to other bacterial proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, super-resolution fluorescence microscopy methods have taken centre stage in microbial cell biology as they can resolve cellular structures well below the diffraction limit (Xiao and Dufrene 2016). One cellular system that has benefitted greatly from these methods is the bacterial cell division machinery or the ‘divisome’. The ‘divisome’ is a complex and transient macromolecular protein assembly containing > 35 different proteins that span all compartments of the cell (Attaibi and den Blaauwen 2022; Haeusser and Margolin 2016). First to arrive at the division site is FtsZ (McQuillen and Xiao 2020), a protein that functions as a molecular foreman that guides the assembly and dynamics of other divisome proteins (den Blaauwen, et al. 2017; Erickson et al. 2010). Due to its broad conservation over many bacterial species, FtsZ could also be a potent target for new antibiotics (Kusuma et al. 2019). As such, an interest in understanding the function of FtsZ has driven the cell division field for the last 25 years (den Blaauwen et al. 2017).
One technical aspect that has hampered studies of FtsZ and other cell division proteins is that reliable labelling is an ongoing challenge for fluorescence microscopy imaging, especially in super-resolution imaging. Considerable progress has been made using N- and C-terminal fusions to fluorescent proteins (FPs), which provide a multi-wavelength tool-box for all modes of fluorescence imaging techniques (Holden 2018; Xiao and Dufrene 2016). And since FPs can be imaged in live cells, temporal changes in localization can be tracked (Holden 2018; Rowlett and Margolin 2014; Strauss et al. 2012). Major drawbacks of FP fusions are that they are large (~ 25 kDa) and that they can perturb the function of the protein they are fused to (as judged by the fact that they cannot always fully complement the phenotype of a deletion strain). As such, they often need to be expressed from plasmids and studied in the context of the native (chromosomally encoded) division protein. An unwanted side-effect of doing this is that the amount of protein in the cell is higher than normal, which can perturb the timing of division and result in an elongated cell phenotype. Eriksson and colleagues elegantly worked around this problem by exploring internal fusion points for FPs in FtsZ (Moore et al. 2017). They noted that all N- and C- terminal fusions are not functional, but that position G55:Q56 could functionally tolerate some but not all FPs. Recently similar approaches have been applied to other division proteins, e.g. FtsN (Lyu et al. 2022) and FtsA (Cameron and Margolin 2023). It should be noted that this approach is extremely time-consuming and not broadly applicable.
An alternative approach is to use fluorescently labelled antibodies to label the native protein. Here, a primary antibody binds to the protein of interest, and a fluorescent dye labelled secondary antibody is used to label the primary antibody. This approach, immunofluorescence microscopy or IFM, has been widely used but is limited by the availability and quality of antisera. Another major drawback here is the large linkage error, defined as the distance from target protein to fluorescent label (Lelek et al. 2021). The linkage error can be in the order of 30–50 nm as antibodies are relatively large and thus of comparable size to the resolving power of various super-resolution microscopy methodologies. This problem is compounded by the fact that most antibodies are polyclonal and bind to multiple epitopes on a protein of interest. These drawbacks create uncertainty about the localization of the protein being studied, which is counter-productive as super-resolution imaging system can have practical resolving powers down to a few tens of nanometres (Carrington et al. 2019). One way of circumventing this issue is to use alternative protein tags, e.g., the HaloTag (Los et al. 2008). These tags are fused to the target protein and a fluorescently labelled substrate (typically an organic dye) is added to bind to the tag. The HaloTag has been successfully used in bacterial cell division studies previously and is especially well suited for single-molecule tracking type experiments (McCausland et al. 2021). One big drawback, however, is that the Halo-tag itself is relatively large (comparable to the size of a FP) and can disrupt the function of the protein of interest.
Herein, we have instead explored the use of nanotags (NTs) and fluorescently labelled nanobodies (NBs) for labelling division proteins in E. coli. Specifically, we have used the ALFA-tag (Gotzke et al. 2019), a small and highly versatile protein tag that can be used for a wide range of biological applications (Akhuli et al. 2022). In bacteriology, the ALFA-tag has thus far been used in super-resolution microscopy of DNA damage repair, as well as in structural biology of cell wall synthesis (Shlosman et al. 2022; Wiktor et al. 2021), but to our knowledge never before in a dynamic live cell setting. We demonstrate that the ALFA-tag can be integrated into the genome of E. coli without affecting the function of the essential cell division protein FtsZ. We further demonstrate that ALFA-labelled FtsZ can be imaged by both STimulated Emission Depletion (STED) microscopy using fluorescently labelled NBs in an immunofluorescence approach, and importantly by expressing fluorescently labelled NBs from plasmids using live cell photoactivated localization microscopy (PALM). The in-vivo approach has a number of advantages over regular FP fusions and immunofluorescence microscopy, which are discussed.
Methods
Strain engineering
The FtsZ-ALFA strain was made using the CRMAGE method (Ronda et al. 2016). In short, the MG1655 strain was transformed with the pMA7CR.2.0 (ampR) and pZS4int-tetR (specR) plasmids, then grown overnight at 37 °C (with shaking) in LB Lennox media containing 100 μg ml−1 ampicillin and 50 μg ml−1 spectinomycin. The culture was back-diluted 1:100 and grown to an OD600 of 0.4–0.5. Production of the λ RED β-protein was induced with 0.2% (w/v) L-arabinose for 15 min. Cells were washed three times in ice-cold water and resuspended in 400 μl of ice-cold water. A 50 μl aliquot of the cells was mixed with 1 μl of 5 μM CRMAGE oligo (Integrated DNA Technologies, USA) and 250 ng of the pMAZ-SK (kanaR) plasmid (containing the sgRNA for the native ftsZ locus) and transferred to an electroporation cuvette with a 1 mm gap. Cells were electroporated at 1.8 kV and then mixed with 950 μl of LB media containing 100 μg/mL ampicillin and 50 μg ml−1 spectinomycin. After a 1-h recovery at 37 °C with shaking, 50 μg ml−1 of kanamycin was added, and cells were incubated for 2 h at 37 °C. The Cas9 protein was induced by the addition of 200 ng ml−1 anhydrotetracycline, and the culture was further incubated at 37 °C (with shaking) overnight. A dilution series was plated on LB agar containing 100 μg ml−1 ampicillin, 50 μg ml−1 spectinomycin and 50 μg ml−1 of kanamycin. Colonies were screened by diagnostic PCR to identify recombinant strains. The sequence of the CRMAGE oligo is listed in Supplementary Table S1. The pMAZ-SK plasmid containing the sgRNA was constructed using the in vivo cloning approach (Bubeck, et al. 1993; Garcia-Nafria et al. 2016; Jones and Howard 1991). In short, the plasmid backbone was amplified by PCR using primers P9 and P10 and the Q5 DNA polymerase (New England Biolabs, USA, NEB #M0491). These primers were obtained from Eurofins Genomics (Germany) and are listed in Supplementary Table S1. The plasmid was sequenced verified by Eurofins Genomics (Germany).
The engineered FtsZ-ALFA strain was subsequently cured from the pMAZ-SK plasmid as previously described with minor modifications (Ronda et al. 2016). In short, cells were grown overnight then washed twice with fresh LB and back diluted in fresh LB with anhydrotetracycline 200 ng ml−1 and 0.2% (w/v) of L-Rhamnose and grown for 24 h. Cultures were plated on Amp/Spect (100 μg ml−1 ampicillin, 50 μg ml−1 spectinomycin) and Kan plates (50 μg ml−1 Kanamycin). The final cured strain MG1655 (FtsZ-ALFA(G55:Q56)) was denoted AB003.
Plasmid construction
The expression plasmid pBAD/HisB-mEos3.2-α-ALFA (kanaR) was constructed using the pBAD/HisB(TIREVOL)-mEos3.2 (ampR) backbone (Addgene plasmid #189,720; (Shilling et al. 2022)). Initially, pBAD/HisB(TIREVOL)-mEos3.2 (ampR) was linearised by PCR using the primers P1 and P2. The coding sequence for the α-ALFA NB was amplified by PCR from the mEGFP-NbALFA plasmid (Addgene plasmid #159,986; (Jin et al. 2021)) using the primers P3 and P4. This PCR product contained 5′ and 3′ extensions (19 and 18 bps, respectively) that overlapped with the desired integration site in the pBAD/HisB(TIREVOL)-mEos3.2 (ampR) backbone so that they could be ligated using the in vivo cloning approach (Bubeck et al. 1993; Garcia-Nafria et al. 2016; Jones and Howard 1991). In short, this required that the PCR products were treated with DpnI enzyme for 60 min at 37 °C, then transformed into MC1061 strain using a standard heat shock protocol. The final clone was called pBAD/HisB-mEos3.2-α-ALFA (ampR). In a second step, the Tn3.17 fragment containing the coding sequence for ß-lactamase (and conferring resistance to ß-lactam antibiotics; ampR) was replaced with the Tn903.1 fragment containing the aminoglycoside 3′ phosphatase (and conferring resistance to aminoglycoside antibiotics; kanaR). pBAD/HisB-mEos3.2-α-ALFA (ampR) was linearised by PCR using the primers P5 and P6. The coding sequence for the Tn903.1 was amplified by PCR from the pET28 (T7pCONS/TIR2)-sfGFP plasmid (Addgene plasmid #154,464;(Shilling et al. 2020)) using the primers P7 and P8. This PCR product contained 5′ and 3′ extensions (15 and 18 bps, respectively) that overlapped with the integration site in the pBAD/HisB-mEos3.2- α -ALFA backbone so that they could be ligated using the in vivo cloning approach (Bubeck et al. 1993; Garcia-Nafria et al. 2016; Jones and Howard 1991). The final clone was called pBAD/HisB-mEos3.2- α -ALFA (kanaR). PCR was carried out with the Q5 DNA polymerase (New England Biolabs, USA, NEB #M0491).
Finally, to construct pBAD/HisB-sfGFP-α-ALFA (kanaR), we exchanged the coding sequence of mEos3.2 for that of sfGFP in the pBAD/HisB-mEos3.2-α -ALFA (kanaR) plasmid using primers P11–P14. The sfGFP coding sequence was PCR amplified from pGI5 plasmid (Iosifidis and Duggin 2020), using primers P13 and P14. The pBAD-mEos3.2-αALFA (kanaR) plasmid backbone was PCR amplified using primers P11 and P12, before combining the PCR products using Gibson assembly using NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, USA, NEB #E2621L). The assembled product was then transformed into E. coli DH5α chemical competent cells before selecting the positive colonies and isolating the plasmid. The isolated pBAD/HisB-sfGFP- α -ALFA (kanaR) plasmid was sequenced to confirm the correct integration of sfGFP coding sequence and transformed into E. coli MG1655 FtsZ-ALFA strain for further analysis.
Primers were obtained from either Eurofins Genomics (Germany) or Integrated DNA Technologies, Inc. and are listed in Supplementary Table S1. Plasmids were sequenced verified by sanger sequencing from Eurofins Genomics (Germany) or Australian Genome Research Facility (AGRF), Sydney.
Bacterial growth and plasmid expression
Pre-cultures of AB003 with or without pBAD/HisB-mEos3.2-α-ALFA were grown overnight in 20 ml of LB with appropriate antibiotics (Ampicillin 100 μg ml−1, Kanamycin 50 μg ml−1) at 30 °C. The following morning the cultures were back-diluted 1:50 in LB and incubated at 30 °C to OD600 0.3 (~ 2 h). AB003 cells without the plasmid were processed for immunofluorescence and STED (see ‘Cell fixation and nanobody labelling for STED’). Cultures of AB003 transformed with pBAD/HisB-mEos3.2-α-ALFA were pelleted and resuspended in fresh M9 minimal media supplemented with 0.2% casamino acids, 1 mM MgSO4, and 0.00005% arabinose (to initiate production mEos3.2-α-ALFA production) and grown at 30 °C for an additional two hours.
Four μl of cell culture were added to an agarose pad and taken for live cell imaging.
Cell fixation and nanobody labelling for STED
Cells were fixed and immunodecorated as described previously (Söderström et al. 2018b). Briefly, 900 μl of ice-cold methanol were added to 100 μl of cell culture (OD600 ~ 0.2–0.5) and incubated for 5 min on ice. Cells were harvested by centrifugation and resuspended in 100 μl of 90% (v/v) ice-cold methanol. For nanobody labelling of cells, roughly 20 μl of cell suspension applied on a cover glass coated with Poly-L-Lysine or on agarose micron-hole beds and left for ~ 2 min before excess culture was removed. After drying, the cells were treated with lysozyme solution (0.2 mg/mL lysozyme, 25 mM Tris–HCl pH 8.0, 50 mM glucose, 10 mM EDTA) for 7–8 min and then rinsed with PBSTS [140 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 0.05% (v/v) Tween 20, 20% (w/v) sucrose], before they were treated with methanol for 1 min and acetone for 1 min and dried. Fixed cells were incubated with 3% (w/v) bovine serum albumin in PBSTS for at least 15 min. Thereafter the cells were washed with PBSTS and incubated with fluorescently labelled nanobodies (i.e. α-ALFA-ATTO488; cat. no. N1502-At488-L, NanoTag Biotechnologies, gift from Dr. Hansjörg Götzke) for 2–3 h at RT. Cells on cover glasses were placed on glass slides with a drop of ProLong Gold antifade as mounting medium and left overnight to harden before imaging, while cells in agarose micron-hole beds were covered with a pre-cleaned cover glass.
Imaging
STED images were acquired on a Leica TCS SP8 STED 3X system, using a HC PL Apo 100 × oil immersion objective with NA 1.40. ATTO488 was excited using a white excitation laser operated at 488 nm, and STED was achieved using a 592 nm laser line. The total intensity of the depletion laser was in the order of 20–500 MW/cm2 for all STED imaging. To achieve time gated STED was a detection time-delay of 0.9–2.5 ns applied. The final pixel size was 13 nm and scanning speed 600 Hz. The pinhole size was varied between 0.4 and 0.9 AU; a smaller pin-hole when imaging cells standing trapped in micron holes and larger when imaging cells lying flat on agarose pads.
Live cell PALM imaging was performed on a Nikon (Ti2-E) N-STORM v5 with NIS v.5.30 using a 100 × 1.49 NA oil objective, operated in quasi-TIRF mode to increase the signal to noise ratio. A stage-top environmental chamber (Oko lab) was used for temperature control, imaging was performed at 37 °C for live cell and time-lapse imaging. Cover glass slides were washed with 80% EtOH, air dried, cleaned for at least 3 min with a plasma cleaner (Harrick plasma, PDC-23G) and used within 15 min of cleaning. Prior to single-molecule acquisition, the green state of mEos3.2 was excited by a 488 nm laser to acquire an epifluorescence image using a FITC emission filter cube. For single-molecule acquisition, mEos3.2 was photoconverted to its red state continuously by a 405 nm laser with increasing working power ranging between 0.1 and 5 W cm−2. As readout, mEos3.2 was excited by a 561 nm laser line operating at an average power between 1 and 2 kW/cm2. The emission was collected by a quad band (Quad405/488/561/647 filter dual cSTORM). The exposure time was 20 ms and ~ 3000–4000 images were typically acquired for each set of images. Drift correction during image acquisition was minimised using the integrated PFS4 (Perfect Focus System). The relatively short PALM image acquisition time (50 Hz) combined with that the overall system drift was consistently less than one half pixel over the series acquisition time, therefore no fiducial markers were needed (Supplementary Fig. 1). Individual images were captured using a sCMOS Flash 4.0 v3 (Hamamatsu) camera using a final pixel size of 32.4 nm.
Image analysis
STED images were deconvoluted using Huygens Professional deconvolution software (SVI, the Netherlands). The resolution in our STED images was ~ 50 nm (Söderström et al. 2018b). Single-molecule raw data were analysed using the Fiji plug-in ThunderSTORM (Ovesny et al. 2014). Super resolution single-molecule images were rendered using Gaussian blur of 20 nm for visualization. Z-ring widths in cells lying down were extracted from fluorescence intensity profile line traces over midcell. A Gaussian distribution was fitted to the intensity profiles in order to extract the Full Width at Half Maximum (FWHM). Radii from cells trapped standing were estimated by manually fitting a circle over the fluorescence traces in Fiji. Statistics and graphs were generated using custom MATLAB scripts, Origin9 Pro and PlotsOfData (Postma and Goedhart 2019).
Cell length measurements
Cells of WT, FtsZ-ALFA, FtsZ-ALFA/mEos3.2-α-ALFA and FtsZ-ALFA/sfGFP-α-ALFA were harvested from growth cultures by centrifugation. An aliquot (2 μl) was placed on an agarose (1.5% w/w) pad and directly imaged under bright-field illumination. Cell lengths of ~ 200 cells from each strain were determined using MicrobeJ (Ducret et al. 2016). Statistics and graphs were generated using custom MATLAB scripts, Origin9 Pro and PlotsOfData (Postma and Goedhart 2019).
Western blotting
Cell extracts from a volume corresponding to 0.2 OD600 units were collected for different strains. The extracts were suspended in loading buffer and resolved by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose membranes using a semi-dry Turbo Transfer-Blot apparatus (Bio-Rad). The membranes were blocked in 5% (w/v) milk and probed with antisera to FtsZ [1:4000] (Agrisera, Sweden).
Results
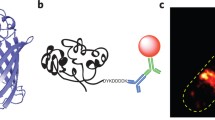
Our goals in this study were (1) to determine whether an essential and normally difficult to tag E. coli cell division protein would remain functional when fused to a NT, and (2) if it could be imaged using super-resolution microscopy. The ALFA-tag is 39 nucleotides long and encodes a 13 amino acid peptide that forms an α-helix (Fig. 1a) (Gotzke et al. 2019). The coding sequence for ALFA was successfully inserted into the chromosomal copy of ftsZ in the E. coli strain MG1655 using CRISPR Optimized MAGE Recombineering (CRMAGE) (Ronda et al. 2016). It was possible to integrate the coding sequence for ALFA in the region of ftsZ corresponding to G55:Q56 by using flanking sequences (GSTLE and LEGST) (Fig. 1b). Attempts to insert the coding sequence for ALFA at the same position (G55:Q56) but without the flanking sequences, or at the C-terminus of FtsZ were not successful, suggesting that the designs were not functionally tolerated (as noted previously with FPs (Moore et al. 2017)). The strain, herein denoted FtsZ-ALFA, had no visible phenotype when visualized by light microscopy (Fig. 1c). The cell lengths were indistinguishable from the wildtype parent strain (Fig. 1e), as was the cell growth (Fig. 1d). Quantitative western blotting of cells harvested in the exponential phase indicated that levels of FtsZ in wild type and the FtsZ-ALFA strain were comparable (Fig. 1f). Taken together, these data indicate that ALFA can be functionally fused to FtsZ in a non-disruptive way. To determine if this tagging strategy is suitable for super resolution imaging, we used two orthogonal labelling approaches: immunofluorescence labelling in fixed cells (Fig. 1g), and plasmid expression of α-ALFA fused to mEos3.2 (a photoconvertible FP) in live cells (Fig. 1h). We then visualised the tagged proteins with two standard super-resolution techniques: STimulated Emission Depletion (STED) microscopy (Hein et al. 2008; Vicidomini et al. 2011) and single-molecule photoactivated localization microscopy (PALM) (Betzig et al. 2006; Greenfield et al. 2009).
Cell viability of the engineered FtsZ-ALFA strain and labelling approaches. a Structure and sequence of the ALFA-tag (Gotzke et al. 2019). b Locus on the chromosome in strain MG1655 where the ALFA-tag is incorporated. c Bright field images of strains used in this study. No morphological differences are noticeable. Scale bars = 5 m. d Growth curves of strains used (n = 3 for each strain). e Cell length measurements. WT = 2.34 ± 0.83 μm, FtsZ-ALFA = 2.61 ± 1.16 μm, FtsZ-ALFA + mEos3.2-α-ALFA = 2.78 ± 1.33 μm (n = 214 for each strain). Mean ± S.D. f Quantitative western blotting indicated that neither the ALFA-tag nor the additional expression of mEos3.2-α-ALFA altered the overall expression of FtsZ to any larger degree. WT = 1,FtsZ-ALFA = 0.99 ± 0.08,FtsZ-ALFA + mEos3.2-α-ALFA = 1.13 ± 0.03. Mean ± S.D. n = 3. Full length WB shown in Supplementary Fig. 3. g Approach 1: immunofluorescence labelling. Cells were fixed, membranes permeabilized followed by labelling with ATTO488 tagged α-ALFA nanobodies recognizing the ALFA-tag. h Approach 2: plasmid expression in live cells. Cells were transformed with a plasmid encoding for mEos3.2-α-ALFA nanobody. In both approaches, FtsZ-ALFA is labelled with fluorescently tagged nanobodies and imaged using super-resolution approaches (Figs. 2 and 4)
STED and immunofluorescence
As a first approach, we used an established immunofluorescence methodology to label FtsZ-ALFA. Cells were fixed with formaldehyde, membranes were permeabilized using standard protocols, and FtsZ-ALFA was labelled with α-ALFA-ATTO488 nanobodies and imaged by confocal fluorescence microscopy and super-resolution STED (Vicidomini et al. 2013). As anticipated, most elongated cells had a visible FtsZ band at the future division site (Fig. 2a) (Sun and Margolin 1998). Images and quantitative measurements of the FWHM indicated that the resolution improvement by STED over conventional confocal imaging was approximately three-fold (Fig. 2b). The average width of the FtsZ-ALFA bands observed from the STED images was determined to be 94 ± 12 nm compared to 243 ± 26 nm for confocal (Fig. 2c). These STED values were slightly lower than those previously reported using STED on IFM labelled FtsZ in E. coli (~ 110 nm) (Söderström et al. 2018b), possibly because the linkage error was reduced using the nanotag approach. FtsZ forms filaments that treadmill around the division septum (Bisson-Filho et al. 2017; Yang et al. 2017). Under the super-resolution light microscope, these FtsZ filaments resemble elongated clusters (Fu et al. 2010; Lyu et al. 2022; Söderström et al. 2018b). Here we visualised FtsZ-ALFA rings in cells trapped in a vertical position using a micron-cage approach (Fig. 2d) (Söderström et al. 2018b). As expected, clearly resolvable FtsZ-ALFA clusters were distributed around the division septum and pseudo-time courses of fixed cells could be constructed from FtsZ-ALFA rings at various stages of constriction, with radii less than 100 nm (Fig. 2e).
Super-resolution STED imaging of FtsZ-ALFA using an α-ALFA nanobody labelled with ATTO488. a Fixed cells immunolabelled and imaged using confocal and STED microscopy. Fluorescence signal overlayed with bright field images. Scale bar = 4 μm. b Longitudinal width (FWHM) of a representative FtsZ ring. Conf = Confocal. c Apparent widths (FWHM) were extracted from line scans as indicated by the white dotted line in the lower cells in the confocal image (A). The mean widths were 243 ± 26 nm (confocal) and 93 ± 12 nm (STED) (n = 54). Boxes indicate S.D., midline is the mean and whiskers incorporate 1-99% of the data range. d Schematic representation of cells lying on an agarose pad and trapped in a vertical position. e STED imaging of FtsZ-ALFA cells labelled with α-ALFA-ATTO488 trapped in a vertical position in micron holes. Representative FtsZ rings of various radii. r indicate radius. Scale bar = 500 nm
Live cell PALM using intracellular expressed fluorescent nanobodies
A limitation of the immunofluorescence approach is that cells are fixed, so it is not possible to follow real dynamics of FtsZ-ALFA over time. To circumvent this problem, we transformed the FtsZ-ALFA strain with engineered plasmids encoding for α-ALFA nanobody fused to sfGFP or the photoconvertible mEos3.2 (a FP compatible with single-molecule imaging). The main benefits of this approach are that (1) we could image and follow live cells throughout division, (2) we could control the amount of α-ALFA (so that not all FtsZ-ALFA molecules were labelled) and (3) we could induce α-ALFA when required. FtsZ-ALFA cells transformed with α-ALFA-GFP (Supplementary Fig. 2) showed localization patterns and dynamics of similar to those of FtsZ-GFP described multiple times previously (Buss et al. 2013; Ma et al. 1996; Mannik et al. 2018; Söderström et al. 2014; Sun and Margolin 1998).
In PALM, the localization of single molecules from stochastically photoswitched or photoactivated fluorescent proteins are determined at sub-pixel resolution in a large series of sequential images from which a composite super-resolution image is created (Betzig, et al. 2006; Coltharp and Xiao 2012; Lelek et al. 2021). After titrating the amount of mEos3.2-α-ALFA, it was possible to identify conditions where our experimental single-molecule approach showed clear FtsZ accumulation at midcell without affecting cell morphology or growth (Figs. 3 and 4a).
Efficient FtsZ-ALFA/mEos3.2-α-ALFA ring detection is concentration dependent. Epifluorescence imaging on FtsZ-ALFA tagged with mEos3.2-α-ALFA in live E. coli cells. mEos3.2 was excited and captured in the green state using a 488-laser line. Images of various conditions with different Arabinose concentrations, ranging from 0.001% to 0%. Too high induction (0.001% and above) produced large bright protein aggregates at the poles, 0.0005% produced too high background, while no induction resulted in essentially no fluorescence signal. As clearly can be seen, under our experimental conditions, 0.00005% Arabinose gave best results. Scale bar = 2 μm
Live cell single-molecule PALM imaging of FtsZ-ALFA using a co-expressed mEos3.2-α-ALFA nanobody. a A typical live cell imaged by bright field optics and using PALM. b Side-by-side comparison of FtsZ-ALFA rings using Epifluorescence and PALM. c Longitudinal width (FWHM) of a representative FtsZ-ALFA ring as determined by fitting a gaussian to fluorescence traces generated drawn over the FtsZ-ALFA rings (e.g., yellow line in (B)). EPI Epifluorescence. d Apparent widths (FWHM) from line scans as indicated by the yellow line in the lower cells in the epifluorescence image (B). The mean widths were 266 ± 22 nm (epifluorescence) and 110 ± 11 nm (PALM) (n = 109). Boxes indicate S.D., midline is the mean and whiskers incorporate 1–95% of the data range. e, Images from a typical time-lapse PALM sequence. Z-ring radius is clearly decreasing over time. Scale bars = 2 μm
Again, quantitative measurements of the FWHM indicated a substantial resolution improvement over diffraction limited imaging (Fig. 4b–c). These Z-rings had an average lateral width of 110 ± 11 nm (n = 109) (Fig. 4c), again similar to previously reported values using live cell PALM on FtsZ-mEos (Coltharp et al. 2016; Fu et al. 2010). One important consideration with this approach is that the expression of mEos3.2-α-ALFA needs to be optimised by titration. If expression is too low, it will not be possible to detect the ALFA nanotag, and if the expression is too high, the background fluorescence will be too high (as the mEos3.2-α-ALFA is fluorescent even when it is not bound to the ALFA nanotag) (Fig. 3) (de Beer and Giepmans 2020). This latter problem is unique to live cell imaging as it is not possible to wash away unbound mEos3.2-α-ALFA (as it is with IFM approaches).
The main advantage of this approach over IFM approaches is arguably that cells are imaged live and thus dynamic processes can be followed in super-resolution and avoid artifacts from fixing cells. To capture FtsZ dynamics at single-molecule resolution, we imaged cells on agarose pads over time. We acquired images every 10 min as it was not feasible to decrease the interval between imaging due to the limited pool of mEos3.2-α-ALFA molecules. Time-lapse imaging showed dynamic FtsZ-rings with decreasing radius over time (Fig. 4d).
Conclusions
In summary, here we have demonstrated that nanotag/nanobody labelling approaches can be used to image bacterial cell division proteins at super resolution. Initially, FtsZ-ALFA rings were visualised in fixed cells using an immunofluorescence microscopy (IFM) protocol and super-resolution STED imaging (see also Söderström et al. 2018a, b; Strauss et al. 2012).
In a second step, we also demonstrated the feasibility of nanobody labelling in live bacteria for the first-time using plasmid encoded α-ALFA nanobodies fused to fluorescent proteins sfGFP or mEos3.2. In the latter, this allowed us to use a single-molecule PALM imaging approach. This approach allowed protein specific cell division dynamics at single-molecule resolution to be monitored over time. The advantages of our live cell nanobody approach over conventional fluorescent protein fusions are two-fold: (1) the ALFA-tags are small and are therefore less likely to interfere with normal protein function, and (2) the generation of the nanobody itself is controllable by induction meaning that its production can be restricted to biologically relevant time-points. Two limitations for this approach are also noted: (1) the expression levels of the nanobody fused to the need to be carefully optimised so that the signal to noise ratio is good, and (2) multicolour experiments are not currently feasible as only one plasmid encoded nanobody (the α-ALFA) is currently available. There are, however, several nanotags (e.g., the SPOT-tag (Virant et al. 2018), so this issue may be resolved in the future.
As a general approach, we believe that nano-tagging can easily be translated to a wide range of other events in bacteria and aid others looking to study time dependent processes at single-molecule resolution in systems otherwise challenging to label using standard approaches.
Data availability
The data supporting the findings of the study are available in this article, or from the corresponding authors upon request.
References
Akhuli D, Dhar A, Viji AS, Bhojappa B, Palani S (2022) ALIBY: ALFA nanobody-based toolkit for imaging and biochemistry in yeast. mSphere 7:e0033322. https://doi.org/10.1128/msphere.00333-22
Attaibi M, den Blaauwen T (2022) An updated model of the divisome: regulation of the septal peptidoglycan synthesis machinery by the divisome. Int J Mol Sci. https://doi.org/10.3390/ijms23073537
Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645. https://doi.org/10.1126/science.1127344
Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, Garner EC (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739–743. https://doi.org/10.1126/science.aak9973
Bubeck P, Winkler M, Bautsch W (1993) Rapid cloning by homologous recombination in vivo. Nucleic Acids Res 21:3601–3602. https://doi.org/10.1093/nar/21.15.3601
Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, Hatem C, Xiao J (2013) In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89:1099–1120. https://doi.org/10.1111/mmi.12331
Cameron TA, Margolin W (2023) Construction and characterization of functional FtsA sandwich fusions for studies of FtsA localization and dynamics during Escherichia coli cell division. J Bacteriol 205:e0037322. https://doi.org/10.1128/jb.00373-22
Carrington G, Tomlinson D, Peckham M (2019) Exploiting nanobodies and Affimers for superresolution imaging in light microscopy. Mol Biol Cell 30:2737–2740. https://doi.org/10.1091/mbc.E18-11-0694
Coltharp C, Xiao J (2012) Superresolution microscopy for microbiology. Cell Microbiol 14:1808–1818. https://doi.org/10.1111/cmi.12024
Coltharp C, Buss J, Plumer TM, Xiao J (2016) Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci USA 113:E1044-1053. https://doi.org/10.1073/pnas.1514296113
de Beer MA, Giepmans BNG (2020) Nanobody-based probes for subcellular protein identification and visualization. Front Cell Neurosci 14:573278. https://doi.org/10.3389/fncel.2020.573278
den Blaauwen T, Hamoen LW, Levin PA (2017) The divisome at 25: the road ahead. Curr Opin Microbiol 36:85–94. https://doi.org/10.1016/j.mib.2017.01.007
Ducret A, Quardokus EM, Brun YV (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. https://doi.org/10.1038/nmicrobiol.2016.77
Erickson HP, Anderson DE, Osawa M (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528. https://doi.org/10.1128/MMBR.00021-10
Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J (2010) In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5:e12682. https://doi.org/10.1371/journal.pone.0012680
Garcia-Nafria J, Watson JF, Greger IH (2016) IVA cloning: A single-tube universal cloning system exploiting bacterial In vivo assembly. Sci Rep 6:27459. https://doi.org/10.1038/srep27459
Gotzke H, Kilisch M, Martinez-Carranza M, Sograte-Idrissi S, Rajavel A, Schlichthaerle T, Engels N, Jungmann R, Stenmark P, Opazo F, Frey S (2019) The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat Commun 10:4403. https://doi.org/10.1038/s41467-019-12301-7
Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J (2009) Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol 7:e1000137. https://doi.org/10.1371/journal.pbio.1000137
Haeusser DP, Margolin W (2016) Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14:305–319. https://doi.org/10.1038/nrmicro.2016.26
Hein B, Willig KI, Hell SW (2008) Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc Natl Acad Sci USA 105:14271–14276. https://doi.org/10.1073/pnas.0807705105
Holden S (2018) Probing the mechanistic principles of bacterial cell division with super-resolution microscopy. Curr Opin Microbiol 43:84–91. https://doi.org/10.1016/j.mib.2017.12.005
Iosifidis G, Duggin IG (2020) Distinct morphological fates of uropathogenic Escherichia coli intracellular bacterial communities: dependency on urine composition and pH. Infect Immun 88: e00884–00819
Jin F, Shen C, Wang Y, Wang M, Sun M, Hattori M (2021) Fluorescence-detection size-exclusion chromatography utilizing nanobody technology for expression screening of membrane proteins. Commun Biol 4:366. https://doi.org/10.1038/s42003-021-01891-y
Jones DH, Howard BH (1991) A rapid method for recombination and site-specific mutagenesis by placing homologous ends on DNA using polymerase chain reaction. Biotechniques 10:62–66
Kusuma KD, Payne M, Ung AT, Bottomley AL, Harry EJ (2019) FtsZ as an antibacterial target: status and guidelines for progressing this avenue. ACS Infect Dis 5:1279–1294. https://doi.org/10.1021/acsinfecdis.9b00055
Lelek M, Gyparaki MT, Beliu G, Schueder F, Griffie J, Manley S, Jungmann R, Sauer M, Lakadamyali M, Zimmer C (2021) Single-molecule localization microscopy. Nat Rev Methods Primers. https://doi.org/10.1038/s43586-021-00038-x
Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV (2008) HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3:373–382. https://doi.org/10.1021/cb800025k
Lyu Z, Yahashiri A, Yang X, McCausland JW, Kaus GM, McQuillen R, Weiss DS, Xiao J (2022) FtsN maintains active septal cell wall synthesis by forming a processive complex with the septum-specific peptidoglycan synthases in E. coli. Nat Commun 13:5751. https://doi.org/10.1038/s41467-022-33404-8
Ma X, Ehrhardt DW, Margolin W (1996) Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA 93:12998–13003
Mannik J, Walker BE, Mannik J (2018) Cell cycle-dependent regulation of FtsZ in Escherichia coli in slow growth conditions. Mol Microbiol 110:1030–1044. https://doi.org/10.1111/mmi.14135
McCausland JW, Yang X, Squyres GR, Lyu Z, Bruce KE, Lamanna MM, Söderström B, Garner EC, Winkler ME, Xiao J, Liu J (2021) Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat Commun 12:609. https://doi.org/10.1038/s41467-020-20873-y
McQuillen R, Xiao J (2020) Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu Rev Biophys 49:309–341. https://doi.org/10.1146/annurev-biophys-121219-081703
Moore DA, Whatley ZN, Joshi CP, Osawa M, Erickson HP (2017) Probing for binding regions of the FtsZ protein surface through site-directed insertions: discovery of fully functional FtsZ-fluorescent proteins. J Bacteriol 199:e00553. https://doi.org/10.1128/JB.00553-16
Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM (2014) ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30:2389–2390. https://doi.org/10.1093/bioinformatics/btu202
Postma M, Goedhart J (2019) PlotsOfData-A web app for visualizing data together with their summaries. PLoS Biol 17:e3000202. https://doi.org/10.1371/journal.pbio.3000202
Ronda C, Pedersen LE, Sommer MO, Nielsen AT (2016) CRMAGE: CRISPR optimized MAGE recombineering. Sci Rep 6:19452. https://doi.org/10.1038/srep19452
Rowlett VW, Margolin W (2014) 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J 107:L17-20. https://doi.org/10.1016/j.bpj.2014.08.024
Shilling PJ, Mirzadeh K, Cumming AJ, Widesheim M, Kock Z, Daley DO (2020) Improved designs for pET expression plasmids increase protein production yield in Escherichia coli. Commun Biol 3:214. https://doi.org/10.1038/s42003-020-0939-8
Shilling PJ, Khananisho D, Cumming AJ, Soderstrom B, Daley DO (2022) Signal amplification of araC pBAD using a standardized translation initiation region. Synth Biol (oxf). https://doi.org/10.1093/synbio/ysac009
Shlosman I, Fivenson EM, Gilman MSA, Sisley TA, Walker S, Bernhardt TG, Kruse AC, Loparo JJ (2022) Allosteric activation of cell wall synthesis during bacterial growth. bioRxiv: https://doi.org/10.1101/2022.11.07.515454
Söderström B, Skoog K, Blom H, Weiss DS, von Heijne G, Daley DO (2014) Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol Microbiol 92:1–9. https://doi.org/10.1111/mmi.12534
Söderström B, Badrutdinov A, Chan H, Skoglund U (2018a) Cell shape-independent FtsZ dynamics in synthetically remodeled bacterial cells. Nat Commun 9:4323. https://doi.org/10.1038/s41467-018-06887-7
Söderström B, Chan H, Shilling PJ, Skoglund U, Daley DO (2018b) Spatial separation of FtsZ and FtsN during cell division. Mol Microbiol 107:387–401. https://doi.org/10.1111/mmi.13888
Strauss MP, Liew ATF, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ (2012) 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biology 10ARTN e1001389 https://doi.org/10.1371/journal.pbio.1001389
Sun Q, Margolin W (1998) FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol 180:2050–2056
Vicidomini G, Moneron G, Han KY, Westphal V, Ta H, Reuss M, Engelhardt J, Eggeling C, Hell SW (2011) Sharper low-power STED nanoscopy by time gating. Nat Methods 8:571–573. https://doi.org/10.1038/nmeth.1624
Vicidomini G, Schonle A, Ta H, Han KY, Moneron G, Eggeling C, Hell SW (2013) STED nanoscopy with time-gated detection: theoretical and experimental aspects. PLoS ONE 8:e54421. https://doi.org/10.1371/journal.pone.0054421
Virant D, Traenkle B, Maier J, Kaiser PD, Bodenhofer M, Schmees C, Vojnovic I, Pisak-Lukats B, Endesfelder U, Rothbauer U (2018) A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat Commun 9:930. https://doi.org/10.1038/s41467-018-03191-2
Wiktor J, Gynna AH, Leroy P, Larsson J, Coceano G, Testa I, Elf J (2021) RecA finds homologous DNA by reduced dimensionality search. Nature 597:426–429. https://doi.org/10.1038/s41586-021-03877-6
Xiao J, Dufrene YF (2016) Optical and force nanoscopy in microbiology. Nat Microbiol 1:16186. https://doi.org/10.1038/nmicrobiol.2016.186
Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355:744–747. https://doi.org/10.1126/science.aak9995
Acknowledgements
The authors would like to thank Alex Toftgaard Nielsen and Morten Nørholm (DTU, Denmark) for plasmids, and help setting up the CRMAGE method. We would also like to thank Dr. Hansjörg Götzke (NanoTag Biotechnologies) for the dye labelled nanobodies. This study was supported by a Japan Society for the Promotion of Science KAKENHI young scientist grant (JP17K15694, Japan), a Chancellor’s PostDoctoral Research Fellow grant (UTS, Australia) and an Australian Research Council Discovery Project grant (DP220101143) to B.S. Work in the SCB unit at OIST is funded by core subsidy from Okinawa Institute of Science and Technology Graduate University. Hannah Brown (UTS) is acknowledged for assistance with the Western blotting. D.O.D. was supported by grants from Novo Nordisk Fonden (0071844) and Carl Trygger stiftelse för Vetenskaplig Forskning (CTS 21:1637).
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
B.S. and D.D. conceptualized the study and designed the experiments. B.S., D.O.D, A.B., E.W. and A.P. performed the experiments. H.C. contributed new reagents. All authors helped with data analysis. B.S and D.O.D. wrote the manuscript with input from all authors. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Westlund, E., Bergenstråle, A., Pokhrel, A. et al. Application of nanotags and nanobodies for live cell single-molecule imaging of the Z-ring in Escherichia coli. Curr Genet 69, 153–163 (2023). https://doi.org/10.1007/s00294-023-01266-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-023-01266-2