Abstract
Cellular systems depend on multiprotein complexes whose functionalities require defined stoichiometries of subunit proteins. Proper stoichiometry is achieved by controlling the amount of protein synthesis and degradation even in the presence of genetic perturbations caused by changes in gene dosage. As a consequence of increased gene copy number, excess subunits unassembled into the complex are synthesized and rapidly degraded by the ubiquitin–proteasome system. This mechanism, called protein-level dosage compensation, is widely observed not only under such perturbed conditions but also in unperturbed physiological cells. Recent studies have shown that recognition of unassembled subunits and their selective degradation are intricately regulated. This review summarizes the nature, strategies, and increasing complexity of protein-level dosage compensation and discusses possible mechanisms for controlling proteome stoichiometry in multiple layers of biological processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cellular systems are exposed to a wide variety of environmental changes leading to fluctuations in gene expression and even cell fate (Elowitz et al. 2002; Raj et al. 2010; Raj and Van Oudenaarden 2008; Raser and O'Shea 2004, 2005). Robust control of intracellular concentrations of gene products for the proper rate of biological processes is a prerequisite for cellular systems. In this context, mechanisms buffering intrinsic and extrinsic perturbations in gene expression are the foundation for maintaining cellular homeostasis and ensure cell survival in response to various environments (Kitano 2007; Masel and Siegal 2009; Stelling et al. 2004). Indeed, recent studies have revealed that in Saccharomyces cerevisiae, transient gene dosage imbalance under physiological conditions, especially during DNA replication (early- and late-replicating genes) and meiosis, can be buffered by adjusting the rates of messenger RNA (mRNA) synthesis and protein degradation, respectively (Bar-Ziv et al. 2016; Eisenberg et al. 2018; Voichek et al. 2016). Furthermore, considering that an increase in the copy number of only a subset of the genome leads to a negative impact on cell growth in S. cerevisiae (Makanae et al. 2013; Sopko et al. 2006), genetic perturbations to a broad range of biological processes must be buffered by various mechanisms.
Genetic perturbations caused by increased gene dosage seem to be mainly buffered post-translationally in yeast and human cells (Dephoure et al. 2014; Stingele et al. 2012). Intracellular protein concentration is maintained at an appropriate level by the balance between protein synthesis and degradation. Genome-wide measurements of mRNA translation by ribosome profiling, which is the deep sequencing of ribosome-protected mRNA fragments (ribosome footprints), revealed that compared to mRNA abundance, ribosome footprint density is better correlated with protein abundance determined by mass spectrometry (Ingolia et al. 2009). Whereas translation efficiency, as measured by ribosome profiling, does not decrease upon an increase in gene dosage as described below, degradation of the resulting excess proteins is accelerated to adjust their concentration. These results indicate that the effects of changes in gene dosage are partially buffered by protein degradation, which is referred to as protein-level dosage compensation. Of note, although dosage compensation at the mRNA level under genetic perturbations is controversial, it has been argued that a feedback control of the mRNA level could occur to reduce protein synthesis (Veitia and Potier 2015).
Protein-level dosage compensation plays a crucial role in fine-tuning stoichiometry of multiprotein complex subunits that compose a large fraction of the proteome (Dephoure et al. 2014; Eisenberg et al. 2018; Ishikawa et al. 2017; Shemorry et al. 2013; Sung et al. 2016), thereby complementing the precisely controlled synthesis of those proteins and maintaining proteome stoichiometry (Ingolia et al. 2018). Although we know that protein degradation mediates the compensation, how unassembled subunits are selectively degraded remains largely an open question. This review presents a current overview of the stoichiometry control system, with the main focus on the nature, functions, and mechanisms of protein-level dosage compensation.
Nature and functions of protein-level dosage compensation
An increase in gene dosage does not necessarily lead to a linear increase in protein level, which is mediated by protein-level dosage compensation that buffers gene dosage effects at the protein level rather than the mRNA level (Fig. 1A). For example, in S. cerevisiae, (i) changes in gene expression by a transient imbalance in gene copy number during meiosis are buffered post-translationally, which is mainly observed for genes encoding multiprotein complex subunits (Eisenberg et al. 2018); (ii) in aneuploid cells (Fig. 1B), the increase in protein levels of genes on an extra chromosome is not necessarily twofold, and these proteins are also enriched in subunits of multiprotein complexes (Dephoure et al. 2014; Torres et al. 2007); (iii) the complex subunits encoded by paralogous genes are likely compensated for modulating protein interactome (Diss et al. 2013, 2017); (iv) genetic screening using multicopy plasmids also revealed that 5 out of 54 tested genes on chromosome I are subject to dosage compensation (Fig. 1A and B) (Ishikawa et al. 2017). These results indicate that protein-level dosage compensation plays a role in buffering gene dosage effects under unperturbed and perturbed conditions.
Protein-level dosage compensation. A Protein-level dosage compensation buffers genetic perturbations post-translationally. B Genetic perturbations caused by increased gene dosage due to aneuploidy and multicopy plasmids. They affect cellular systems differently (Bonney et al. 2015), and not all of the compensated proteins identified in these conditions overlap (Ishikawa et al. 2017)
Systematic identification of the compensated proteins revealed that around 10–20% and 25% of the genome and 70% and 57% of genes encoding subunits are subject to dosage compensation in yeast and human cells, respectively (Dephoure et al. 2014; Ishikawa et al. 2017; Stingele et al. 2012). This finding is consistent with the ‘balance hypothesis’ that links stoichiometric imbalance of the complex subunits and a negative impact on cell growth (Papp et al. 2003; Veitia et al. 2008). Indeed, overexpression experiments have confirmed this hypothesis in 13 out of 49 tested subunit pairs (27%) (Makanae et al. 2013). It is unclear whether the other 36 complexes are compensated, thereby avoiding growth defects, but recent results suggest that dosage compensation contributes to cellular robustness (Ascencio et al. 2021). The effect of gene duplication on fitness was measured for 899 essential genes in budding yeast, and it was found that only about 10% of the genes were associated with fitness disadvantage and that the effect on fitness was limited because genes encoding subunits were protected by dosage compensation (Ascencio et al. 2021). Therefore, it is concluded that the primary function of protein-level dosage compensation is to fine-tune stoichiometry of multiprotein complex subunits.
Since not all subunits are subject to dosage compensation, there may be conditions that determine their potential to be substrates. The assembly interface of the unassembled subunit is free from the partner subunit, and the number of direct interactions with partner subunits should reflect how much of the surface area of unassembled subunit is occupied by non-interacting parts. Theoretically, subunits with a higher number of interactions are more unstable when unassembled than subunits with fewer interactions (Veitia et al. 2008). Thus, the number of interactions seems to be one factor in determining the substrate of dosage compensation, which is supported experimentally (Ishikawa et al. 2017; Mueller et al. 2015).
Mechanisms of protein-level dosage compensation
Ribosome profiling experiments have shown that there is a high correlation between translation efficiency of mRNAs encoding complex subunits and their stoichiometry (Li et al. 2014). This phenomenon ‘proportional synthesis strategy’ has been observed in bacteria, yeast, zebrafish, mouse, and human cells, indicating that regulation of protein synthesis generally contributes to stoichiometry control under physiological conditions (Taggart and Li 2018). However, proportional synthesis is not always sufficient for proper stoichiometry because translation efficiency is matched within roughly 20% and this difference is considerably large, especially for highly expressed proteins (Li et al. 2014). For example, in yeast data set containing well-characterized complexes, two out of six subunits of the signal recognition particle are synthesized in excess. Intriguingly, ribosome profiling has revealed that translation efficiency of mRNAs encoding subunits of multiprotein complexes does not decrease with increasing gene dosage, excess proteins are synthesized with normal translation efficiency, and they are rapidly degraded by the ubiquitin–proteasome system (Fig. 1A) (Dephoure et al. 2014; Eisenberg et al. 2018; Ishikawa et al. 2017; Taggart and Li 2018; Thorburn et al. 2013).
The half-life of physiologically over-synthesized subunits tends to be shorter than that of proportionally synthesized subunits (Taggart and Li 2018), consistent with several studies showing that degradation of unassembled subunits by the ubiquitin–proteasome system is the compensation mechanism (Figs. 1A and 2). First, the compensation is generally reduced in the presence of the proteasome inhibitor MG132 (Dephoure et al. 2014). Second, polyubiquitinated forms of the compensated proteins accumulate in the proteasome-deficient mutant (Ishikawa et al. 2017). Third, yeast E3 ubiquitin ligases Doa10, Not4, and Tom1 and a mammalian E2 ubiquitin-conjugating/E3 ligase hybrid protein UBE2O were identified to be responsible for stoichiometry control (Hwang et al. 2010; Shemorry et al. 2013; Sung et al. 2016; Yanagitani et al. 2017). Multiple E3 ligases (Not4 and Tom1) are rarely involved in the compensation of the same subunits (Ishikawa et al. 2020). Identifying such combinations and other compensation E3 ligases is particularly promising for a better understanding of the compensation mechanisms. Likewise, because nuclear-localized E3 ligase Tom1 tends to compensate nuclear proteins (e.g., histone H2A/H3/H4, ribosomal proteins, and the RNase P/MRP subunits) (Ishikawa et al. 2020; Singh et al. 2009; Sung et al. 2016), it is also important to determine the substrate preference of the compensation E3 ligases.
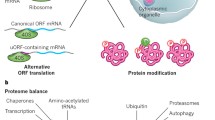
Multilayered structure of the stoichiometry control system. The first layer is the proportional synthesis strategy that enables scaling between stoichiometry and translation efficiency. An exemplary protein complex with 1:2 stoichiometry is shown. If translation efficiency is not proportional to stoichiometry of the complex, or if excess subunits are synthesized due to increased gene dosage, the next layers correct stoichiometric imbalance. The second layer is to control the amount of (sub)complex, thereby stabilizing the partner subunits. The third layer is the rapid degradation of excess subunits by the ubiquitin–proteasome system
Aneuploid cells are prone to form protein aggregates (Oromendia et al. 2012), which is mainly due to stoichiometric imbalance of protein complexes (Brennan et al. 2019). This study has also revealed that both aggregate formation and proteasomal degradation are involved in the mechanism of stoichiometry control and that dosage compensation of the same protein by both pathways rarely occurs (Brennan et al. 2019). Aggregation of excess subunits may protect cells from gene dosage effects that could cause inappropriate protein-protein interactions (Levy et al. 2012). However, the fate of aggregated excess subunits is unclear, and it remains to be investigated whether they are eventually degraded. The degradation of such large aggregates is unlikely to be mediated by the ubiquitin–proteasome pathway; rather, selective autophagy seems to play more important roles in aggregate clearance (Tyedmers et al. 2010). Although p62-mediated autophagy seems to be active in aneuploid human cells (Stingele et al. 2012, 2013), there is no direct evidence for selective degradation of unassembled subunits through this pathway. Because mouse but not yeast aneuploid cells are sensitive to the autophagy inhibitor chloroquine (Tang et al. 2011; Torres et al. 2007), it might be possible that autophagy-mediated dosage compensation occurs specifically in aneuploid mammalian cells.
Another point to consider is the fact that the stability of a subunit protein is affected by its partner subunit; the amount of the Rbg1–Tma46 heterodimer changes in response to the amount of TMA46 (Ishikawa et al. 2017). This regulation may occur also at the subcomplex level, as (i) the analysis of the Pop6–Pop7 subcomplex of the RNase P/MRP complexes has shown that overexpression of Pop6 stabilizes Pop7 and vice versa (Ishikawa et al. 2020), and (ii) deletion of the gene encoding oligosaccharyltransferase (OST) complex subunit destabilizes other subunits of the same OST subcomplex (Mueller et al. 2015). These results suggest that stoichiometry is controlled at the subcomplex level before multiple subcomplexes are assembled into a fully formed multiprotein complex, providing insight into the layered structure of the stoichiometry control system (Fig. 2). Whether these extra subcomplexes will eventually be degraded or assembled into functional complexes remains to be investigated, but the stoichiometric imbalance could be fine-tuned by changing the amount of subcomplexes as well as their degradation by the proteasome. Indeed, degradation rates of subunits belonging to the same subcomplex of the OST complex influence each other (Mueller et al. 2015). This observation has suggested that stoichiometry of this complex is fine-tuned by both the second and third layers (Fig. 2) and that the stability of the subunit corresponds to the state of assembly: (stable) fully assembled complex ≥ subcomplex > unassembled monomeric subunit (unstable). It is further supported by the observation of a similar degree of dosage compensation between subunits of the same subcomplex of the 26S proteasome (Ascencio et al. 2021). Notably, the second layer does not apply to dosage-sensitive complexes including tubulin, whose excess is associated with toxicity, and thus its stoichiometry is tightly controlled in the first layer (Katz et al. 1990; Li et al. 2014; Makanae et al. 2013).
Recognition of unassembled subunits for selective degradation and assembly control
While evidence for degradation-mediated stoichiometry control is accumulating as described above, how cells recognize and selectively degrade unassembled subunits is not well understood. The Ac/N-end rule pathway is one proposed mechanism by which N-terminal acetylation of proteins acts as a degradation signal (N-degron) that could be recognized by the compensation E3 ligases, Not4 and Doa10 (N-recognin) (Fig. 3A) (Hwang et al. 2010; Shemorry et al. 2013; Varshavsky 2011). Dosage compensation by this pathway is based on the assumption that N-degron of assembled subunits is shielded and inaccessible by N-recognin when in complex with the other subunits in the same complex, while that of unassembled subunits is exposed and accessible by N-recognin.
Possible mechanisms that regulate proteome stoichiometry and intracellular protein concentration. A Selective degradation of unassembled subunits by the Ac/N-end rule pathway. The N-degron of assembled subunits is shielded by partner subunits of the same complex and is not targeted by N-recognin, while that of unassembled subunits is accessible and enables selective degradation. B A ‘lysine hide-and-seek’ model for co-translational dosage compensation that degrades nascent polypeptides emerging from ribosomes translating mRNAs encoding excess subunits. This model is based on the assumption that in the absence of a co-translationally assembled partner subunit, lysine residues at the assembly interface of nascent polypeptides are ubiquitinated and subsequently degraded by the proteasome. C A model for regulating intracellular protein concentration by controlling cell size. When the increase in protein concentration exceeds the degradation capacity (e.g., proteasome overload), protein concentration is diluted and proteome stoichiometry is maintained constant by increasing the cell size
N-terminal acetylation is a widespread protein modification mediated by N-acetyltransferases (NATs), accounting for about 68% and 85% of the yeast and human proteomes, respectively (Van Damme et al. 2011). In yeast cells, there are five NATs (NatA–NatE) bound to the ribosome, and in principle each NAT acetylates specific N-terminal residues, which has been reviewed extensively (Aksnes et al. 2016, 2019; Starheim et al. 2012). N-acetylation is found in the majority of proteomes, but the extent to which NATs-mediated compensation is widespread and contributes to proper stoichiometry has not been investigated. Recently, we examined whether the compensated proteins are stabilized in the absence of the catalytic subunit (Naa10–Naa50) of each NAT and found that the contribution of NATs to the compensation varies among subunits (90% for 2 and 10–40% for 12 out of 14 subunits) (Ishikawa et al. 2020). These two proteins, Pop3 and Bet4, were almost fully uncompensated in naa40∆ cells, although only histone H2A/H4 are known as substrates of N-acetylation by Naa40 (Song et al. 2003). These histone subunits were not stabilized in naa40∆ but less compensated in tom1∆ cells, suggesting NATs-independent stoichiometry control (Ishikawa et al. 2020). In contrast, Rmp1 and Bet4 were compensated via multiple NATs; Rmp1 was compensated to a lesser extent in the absence of Naa20, which is responsible for its N-acetylation, compared to Naa10. A comprehensive analysis of dosage compensation using mutants lacking multiple NATs is needed to investigate their functional interactions and working principles in the stoichiometry control system. We also found that NATs-mediated compensation becomes relevant when its target protein is overexpressed, consistent with the observation that N-acetylation rarely acts as a degron under physiological conditions (Kats et al. 2018). These results suggest that the Ac/N-end rule pathway is partially responsible for stoichiometry control.
Several different strategies exist for controlling the assembly of proteins localized to the endoplasmic reticulum (ER). The unassembled subunit of the Na/K-ATPase in Xenopus oocyte is retained in the ER and associates with the ER chaperone BiP until it is complexed with partner subunits (Beggah et al. 1996). The binding of BiP reduces the degradation of unassembled subunits of this complex and increases its stability. Another strategy is to control the assembly interface between subunits. Recently, a ubiquitously expressed kinase WNK1 has been identified as an assembly factor for the ER membrane complex (EMC) in human cells (Pleiner et al. 2021). This study has revealed that WNK1 selectively binds to unassembled EMC2 and competes with the binding of E3 ubiquitin ligases (MKRN1 and HUWE1) to protect it from degradation. WNK1 seems to dissociate from EMC2 when the partner subunit is available for assembly (Pleiner et al. 2021), which may be another strategy in the second layer of the stoichiometry control system (Fig. 2). Thus, identifying factors responsible for this type of assembly control will also facilitate an understanding of how cells regulate proteome stoichiometry.
Perspectives
An important issue arising from previous studies is quality control of excess subunits at the co-translational level. One plausible model is that failures in complex assembly are monitored co-translationally by assembly factors that bind to nascent polypeptides emerging from the ribosome, as proposed in (Juszkiewicz and Hegde 2018). On the other hand, co-translational degradation of nascent polypeptides with N-degron in the Arg/N-end rule pathway (Turner and Varshavsky 2000) raises another possibility that protein-level dosage compensation occurs even during the synthesis of unassembled subunits. Co-translationally ubiquitinated nascent polypeptides undergo proteasomal degradation, which is present in yeast and mammalian cells, but its functions remain unclear (Duttler et al. 2013; Wang et al. 2013). Given that cells can recognize whether the assembly interface of nascent subunit emerging from the ribosome is shielded by a partner subunit, one possible mechanism for co-translational dosage compensation might be ubiquitination of lysine residues at the unpartnered assembly interface of excess subunits (Fig. 3B). Controlling the complex stoichiometry by this mechanism would be cost-beneficial because the absence of one subunit leads to aggregation of a co-translationally assembled partner subunit (Shiber et al. 2018). The compensation E3 ligase Not4 binds to translating ribosomes (Dimitrova et al. 2009; Halter et al. 2014; Preissler et al. 2015); however, deletion of Not4 doubles the amount of co-translationally ubiquitinated nascent polypeptides, suggesting that Not4 may not contribute to co-translational ubiquitination (Duttler et al. 2013). Co-translational ubiquitination is partially reduced in the absence of Hul5, Hrd1, or both Hel2 and Ltn1 (up to 25% in hel2∆ ltn1∆ cells) (Duttler et al. 2013). This reduction in the double mutant seems to be due to the lack of ribosome-associated quality control, which is required for degradation of aberrant nascent polypeptides by the ubiquitin–proteasome system (Brandman and Hegde 2016; Joazeiro 2019), but the functional impact of the other fraction of co-translational ubiquitination remains an important question.
While many studies have shown that dosage compensation mainly targets subunits of multiprotein complexes as described above, evidence is accumulating for the global control of intracellular protein concentration by the regulation of cell size (Fig. 3C). Examples include: (i) overexpression of a positive regulator of mammalian cell size, called Largen, promotes translation of mRNAs, including those encoding histone and mitochondrial proteins (Yamamoto et al. 2014); (ii) overexpression of unneeded exogenous protein (mCherry) results in an increase in cell volume as well as endogenous protein levels in S. cerevisiae (Kafri et al. 2016); (iii) aneuploidy leads to a larger cell volume, which seems to be correlated with increased protein expression due to an extra chromosome (Thorburn et al. 2013); and (iv) cyanobacteria maintain constant protein concentration when genome copy number is increased, which coincides with an increase in cell volume (Zheng and O'Shea 2017). Because proteotoxicity is thought to result from an overload of degradation machinery (Harper and Bennett 2016; Moriya 2015; Oromendia and Amon 2014), perhaps it is more advantageous to regulate cell size than to enhance degradation to survive under such challenging conditions. Therefore, a quantitative study of the relationship between mRNA translation and protein degradation would provide insight into the mechanism by which protein concentration is optimized by regulating cell size.
References
Aksnes H, Drazic A, Marie M, Arnesen T (2016) First things first: vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci 41:746–760. https://doi.org/10.1016/j.tibs.2016.07.005
Aksnes H, Ree R, Arnesen T (2019) Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol Cell 73:1097–1114. https://doi.org/10.1016/j.molcel.2019.02.007
Ascencio D, Diss G, Gagnon-Arsenault I, Dubé AK, Deluna A, Landry CR (2021) Expression attenuation as a mechanism of robustness against gene duplication. Proc Natl Acad Sci 118:e2014345118. https://doi.org/10.1073/pnas.2014345118
Bar-Ziv R, Voichek Y, Barkai N (2016) Dealing with gene-dosage Imbalance during S phase. Trends Genet 32:717–723. https://doi.org/10.1016/j.tig.2016.08.006
Beggah A, Mathews P, Beguin P, Geering K (1996) Degradation and endoplasmic reticulum retention of unassembled α- and β-Subunits of Na, K-ATPase correlate with interaction of BiP. J Biol Chem 271:20895–20902. https://doi.org/10.1074/jbc.271.34.20895
Bonney ME, Moriya H, Amon A (2015) Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Genes Dev 29:898–903. https://doi.org/10.1101/gad.261743.115
Brandman O, Hegde RS (2016) Ribosome-associated protein quality control. Nat Struct Mol Biol 23:7–15. https://doi.org/10.1038/nsmb.3147
Brennan CM, Vaites LP, Wells JN, Santaguida S, Paulo JA, Storchova Z, Harper JW, Marsh JA, Amon A (2019) Protein aggregation mediates stoichiometry of protein complexes in aneuploid cells. Genes Dev 33:1031–1047. https://doi.org/10.1101/gad.327494.119
Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM (2014) Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. https://doi.org/10.7554/elife.03023
Dimitrova LN, Kuroha K, Tatematsu T, Inada T (2009) Nascent peptide-dependent translation arrest leads to not4p-mediated protein degradation by the proteasome. J Biol Chem 284:10343–10352. https://doi.org/10.1074/jbc.m808840200
Diss G, Dubé AK, Boutin J, Gagnon-Arsenault I, Landry CR (2013) A systematic approach for the genetic dissection of protein complexes in living cells. Cell Rep 3:2155–2167. https://doi.org/10.1016/j.celrep.2013.05.004
Diss G, Gagnon-Arsenault I, Dion-Coté AM, Vignaud H, Ascencio DI, Berger CM, Landry CR (2017) Gene duplication can impart fragility, not robustness, in the yeast protein interaction network. Science 355:630–634. https://doi.org/10.1126/science.aai7685
Duttler S, Pechmann S, Frydman J (2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell 50:379–393. https://doi.org/10.1016/j.molcel.2013.03.010
Eisenberg AR, Higdon A, Keskin A, Hodapp S, Jovanovic M, Brar GA (2018) Precise post-translational tuning occurs for most protein complex components during meiosis. Cell Rep 25:3603-3617.e3602. https://doi.org/10.1016/j.celrep.2018.12.008
Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297:1183–1186. https://doi.org/10.1126/science.1070919
Halter D, Collart MA, Panasenko OO (2014) The Not4 E3 Ligase and CCR4 deadenylase play distinct roles in protein quality control. PLoS One 9:e86218. https://doi.org/10.1371/journal.pone.0086218
Harper JW, Bennett EJ (2016) Proteome complexity and the forces that drive proteome imbalance. Nature 537:328–338. https://doi.org/10.1038/nature19947
Hwang CS, Shemorry A, Varshavsky A (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327:973–977. https://doi.org/10.1126/science.1183147
Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. https://doi.org/10.1126/science.1168978
Ingolia NT, Hussmann JA, Weissman JS (2018) Ribosome profiling: global views of translation. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a032698
Ishikawa K, Makanae K, Iwasaki S, Ingolia NT, Moriya H (2017) Post-translational dosage compensation buffers genetic perturbations to stoichiometry of protein complexes. PLoS Genet 13:e1006554. https://doi.org/10.1371/journal.pgen.1006554
Ishikawa K, Ishihara A, Moriya H (2020) Exploring the complexity of protein-level dosage compensation that fine-tunes stoichiometry of multiprotein complexes. PLoS Genet 16:e1009091. https://doi.org/10.1371/journal.pgen.1009091
Joazeiro CAP (2019) Mechanisms and functions of ribosome-associated protein quality control. Nat Rev Mol Cell Biol 20:368–383. https://doi.org/10.1038/s41580-019-0118-2
Juszkiewicz S, Hegde RS (2018) Quality control of orphaned proteins. Mol Cell 71:443–457. https://doi.org/10.1016/j.molcel.2018.07.001
Kafri M, Metzl-Raz E, Jona G, Barkai N (2016) The cost of protein production. Cell Rep 14:22–31. https://doi.org/10.1016/j.celrep.2015.12.015
Kats I, Khmelinskii A, Kschonsak M, Huber F, Kniess RA, Bartosik A, Knop M (2018) Mapping degradation signals and pathways in a eukaryotic N-terminome. Mol Cell 70(488–501):e485. https://doi.org/10.1016/j.molcel.2018.03.033
Katz W, Weinstein B, Solomon F (1990) Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol 10:5286–5294. https://doi.org/10.1128/mcb.10.10.5286-5294.1990
Kitano H (2007) Towards a theory of biological robustness. Mol Syst Biol 3:137. https://doi.org/10.1038/msb4100179
Levy ED, De S, Teichmann SA (2012) Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci 109:20461–20466. https://doi.org/10.1073/pnas.1209312109
Li GW, Burkhardt D, Gross C, Weissman JS (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. https://doi.org/10.1016/j.cell.2014.02.033
Makanae K, Kintaka R, Makino T, Kitano H, Moriya H (2013) Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res 23:300–311. https://doi.org/10.1101/gr.146662.112
Masel J, Siegal ML (2009) Robustness: mechanisms and consequences. Trends Genet 25:395–403. https://doi.org/10.1016/j.tig.2009.07.005
Moriya H (2015) Quantitative nature of overexpression experiments. Mol Biol Cell 26:3932–3939. https://doi.org/10.1091/mbc.e15-07-0512
Mueller S, Wahlander A, Selevsek N, Otto C, Ngwa EM, Poljak K, Frey AD, Aebi M, Gauss R (2015) Protein degradation corrects for imbalanced subunit stoichiometry in OST complex assembly. Mol Biol Cell 26:2596–2608. https://doi.org/10.1091/mbc.e15-03-0168
Oromendia AB, Amon A (2014) Aneuploidy: implications for protein homeostasis and disease. Dis Model Mech 7:15–20. https://doi.org/10.1242/dmm.013391
Oromendia AB, Dodgson SE, Amon A (2012) Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26:2696–2708. https://doi.org/10.1101/gad.207407.112
Papp B, Pál C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424:194–197. https://doi.org/10.1038/nature01771
Pleiner T, Hazu M, Tomaleri GP, Januszyk K, Oania RS, Sweredoski MJ, Moradian A, Guna A, Voorhees RM (2021) WNK1 is an assembly factor for the human ER membrane protein complex. Mol Cell. https://doi.org/10.1016/j.molcel.2021.04.013
Preissler S, Reuther J, Koch M, Scior A, Bruderek M, Frickey T, Deuerling E (2015) Not4-dependent translational repression is important for cellular protein homeostasis in yeast. EMBO J 34:1905–1924. https://doi.org/10.15252/embj.201490194
Raj A, Van Oudenaarden A (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135:216–226. https://doi.org/10.1016/j.cell.2008.09.050
Raj A, Rifkin SA, Andersen E, Van Oudenaarden A (2010) Variability in gene expression underlies incomplete penetrance. Nature 463:913–918. https://doi.org/10.1038/nature08781
Raser JM, O’Shea EK (2004) Control of stochasticity in eukaryotic gene expression. Science 304:1811–1814. https://doi.org/10.1126/science.1098641
Raser JM, O’Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309:2010–2013. https://doi.org/10.1126/science.1105891
Shemorry A, Hwang CS, Varshavsky A (2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell 50:540–551. https://doi.org/10.1016/j.molcel.2013.03.018
Shiber A, Döring K, Friedrich U, Klann K, Merker D, Zedan M, Tippmann F, Kramer G, Bukau B (2018) Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561:268–272. https://doi.org/10.1038/s41586-018-0462-y
Singh RK, Kabbaj MH, Paik J, Gunjan A (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol 11:925–933. https://doi.org/10.1038/ncb1903
Song OK, Wang X, Waterborg JH, Sternglanz R (2003) An Nalpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J Biol Chem 278:38109–38112. https://doi.org/10.1074/jbc.C300355200
Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, Boone C, Andrews B (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21:319–330. https://doi.org/10.1016/j.molcel.2005.12.011
Starheim KK, Gevaert K, Arnesen T (2012) Protein N-terminal acetyltransferases: when the start matters. Trends Biochem Sci 37:152–161. https://doi.org/10.1016/j.tibs.2012.02.003
Stelling J, Sauer U, Szallasi Z, Doyle FJ, Doyle J (2004) Robustness of cellular functions. Cell 118:675–685. https://doi.org/10.1016/j.cell.2004.09.008
Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z (2012) Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8:608. https://doi.org/10.1038/msb.2012.40
Stingele S, Stoehr G, Storchova Z (2013) Activation of autophagy in cells with abnormal karyotype. Autophagy 9:246–248. https://doi.org/10.4161/auto.22558
Sung MK, Porras-Yakushi TR, Reitsma JM, Huber FM, Sweredoski MJ, Hoelz A, Hess S, Deshaies RJ (2016) A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. https://doi.org/10.7554/eLife.19105
Taggart JC, Li GW (2018) Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst 7:580-589.e584. https://doi.org/10.1016/j.cels.2018.11.003
Tang YC, Williams BR, Siegel JJ, Amon A (2011) Identification of aneuploidy-selective antiproliferation compounds. Cell 144:499–512. https://doi.org/10.1016/j.cell.2011.01.017
Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A (2013) Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell 24:1274–1289. https://doi.org/10.1091/mbc.e12-07-0520
Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. https://doi.org/10.1126/science.1142210
Turner GC, Varshavsky A (2000) Detecting and measuring cotranslational protein degradation in vivo. Science 289:2117–2120. https://doi.org/10.1126/science.289.5487.2117
Tyedmers J, Mogk A, Bukau B (2010) Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11:777–788. https://doi.org/10.1038/nrm2993
Van Damme P, Hole K, Pimenta-Marques A, Helsens K, Vandekerckhove J, Martinho RG, Gevaert K, Arnesen T (2011) NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet 7:e1002169. https://doi.org/10.1371/journal.pgen.1002169
Varshavsky A (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20:1298–1345. https://doi.org/10.1002/pro.666
Veitia RA, Potier MC (2015) Gene dosage imbalances: action, reaction, and models. Trends Biochem Sci 40:309–317. https://doi.org/10.1016/j.tibs.2015.03.011
Veitia RA, Bottani S, Birchler JA (2008) Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet 24:390–397. https://doi.org/10.1016/j.tig.2008.05.005
Voichek Y, Bar-Ziv R, Barkai N (2016) Expression homeostasis during DNA replication. Science 351:1087–1090. https://doi.org/10.1126/science.aad1162
Wang F, Durfee LA, Huibregtse JM (2013) A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell 50:368–378. https://doi.org/10.1016/j.molcel.2013.03.009
Yamamoto K, Gandin V, Sasaki M, McCracken S, Li W, Jennifer A, Wang F, Wakutani Y, Alexandrova R, Yathor MPJ, Inoue S, Itsumi M, Lapin V, Haight J, Wakeham A, Shahinian A, Ikura M, Topisirovic I, Sonenberg N, Mak TW (2014) Largen: a molecular regulator of mammalian cell size control. Mol Cell 53:904–915. https://doi.org/10.1016/j.molcel.2014.02.028
Yanagitani K, Juszkiewicz S, Hegde RS (2017) UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357:472–475. https://doi.org/10.1126/science.aan0178
Zheng XY, O’Shea EK (2017) Cyanobacteria maintain constant protein concentration despite genome copy-number variation. Cell Rep 19:497–504. https://doi.org/10.1016/j.celrep.2017.03.067
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science: Grant-in-Aid for JSPS Research Fellow (16J00852) and JSPS Overseas Research Fellowships (202160021). I apologize for not citing numerous papers that contribute to this topic. Finally, I would like to express my sincere thanks to Angelika Amon (1967–2020) and her team for their important work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Michael Polymenis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ishikawa, K. Multilayered regulation of proteome stoichiometry. Curr Genet 67, 883–890 (2021). https://doi.org/10.1007/s00294-021-01205-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-021-01205-z