Abstract
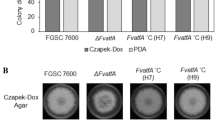
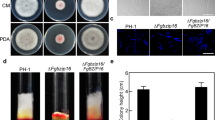
Fumonisins are a group of mycotoxins produced by maize pathogen Fusarium verticillioides that pose health concerns to humans and animals. Yet we still lack a clear understanding of the mechanism of fumonisins regulation during pathogenesis. The heterotrimeric G protein complex, which consists of canonical subunits and various regulators of G-protein signaling (RGS) proteins, plays an important role in transducing signals under environmental stress. Earlier studies demonstrated that Gα and Gβ subunits are positive regulators of fumonisin B1 (FB1) biosynthesis and that two RGS genes, FvFlbA1 and FvFlbA2, were highly upregulated in Gβ deletion mutant ∆Fvgbb1. Notably, FvFlbA2 has a negative role in FB1 regulation. While many fungi contain a single copy of FlbA, F. verticillioides harbors two putative FvFlbA paralogs, FvFlbA1 and FvFlbA2. In this study, we further characterized functional roles of FvFlbA1 and FvFlbA2. While ∆FvflbA1 deletion mutant exhibited no significant defects, ∆FvflbA2 and ∆FvflbA2/A1 mutants showed thinner aerial hyphal growth while promoting FB1 production. FvFlbA2 is required for proper expression of key conidia regulation genes, including putative FvBRLA, FvWETA, and FvABAA, while suppressing FUM21, FUM1, and FUM8 expression. Split luciferase assays determined that FvFlbA paralogs interact with key heterotrimeric G protein components, which in turn will lead altered G-protein-mediated signaling pathways that regulate FB1 production and asexual development in F. verticillioides.








Similar content being viewed by others
References
Aerts D, Hauer EE, Ohm RA, Arentshorst M, Teertstra WR, Phippen C, Ram AFJ, Frisvad JC, Wosten HAB (2018) The FlbA-regulated predicted transcription factor Fum21 of Aspergillus niger is involved in fumonisin production. Antonie Van Leeuwenhoek 111:311–322. https://doi.org/10.1007/s10482-017-0952-1
Alexander NJ, Proctor RH, McCormick SP (2009) Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev 28:198–215. https://doi.org/10.1080/15569540903092142
Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J (2006) DEP-domain-mediated regulation of GPCR signaling responses. Cell 126:1079–1093. https://doi.org/10.1016/j.cell.2006.07.030
Blacutt AA, Gold SE, Voss KA, Gao M, Glenn AE (2018) Fusarium verticillioides: advancements in understanding the toxicity, virulence, and niche adaptations of a model mycotoxigenic pathogen of maize. Phytopathology 108:312–326. https://doi.org/10.1094/PHYTO-06-17-0203-RVW
Brown DW, Butchko RA, Busman M, Proctor RH (2007) The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryot Cell 6:1210–1218. https://doi.org/10.1128/EC.00400-06
Burchett SA, Flanary P, Aston C, Jiang L, Young KH, Uetz P, Fields S, Dohlman HG (2002) Regulation of stress response signaling by the N-terminal dishevelled/EGL-10/pleckstrin domain of Sst2, a regulator of G protein signaling in Saccharomyces cerevisiae. J Biol Chem 277:22156–22167. https://doi.org/10.1074/jbc.M202254200
Christensen S, Borrego E, Shim WB, Isakeit T, Kolomiets M (2012) Quantification of fungal colonization, sporogenesis, and production of mycotoxins using kernel bioassays. J Vis Exp. https://doi.org/10.3791/3727
Desjardins AE, Plattner RD (2000) Fumonisin B(1)-nonproducing strains of Fusarium verticillioides cause maize (Zea mays) ear infection and ear rot. J Agric Food Chem 48:5773–5780. https://doi.org/10.1021/jf000619k
Desjardins AE, Munkvold GP, Plattner RD, Proctor RH (2002) FUM1–a gene required for fumonisin biosynthesis but not for maize ear rot and ear infection by Gibberella moniliformis in field tests. Mol Plant Microbe Interact 15:1157–1164. https://doi.org/10.1094/MPMI.2002.15.11.1157
Dohlman HG (2009) RGS proteins the early days. Prog Mol Biol Transl Sci 86:1–14. https://doi.org/10.1016/S1877-1173(09)86001-8
Dohlman HG, Song JP, Apanovitch DM, DiBello PR, Gillen KM (1998) Regulation of G protein signalling in yeast. Semin Cell Dev Biol 9:135–141. https://doi.org/10.1006/scdb.1998.0218
Hicks JK, Yu JH, Keller NP, Adams TH (1997) Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J 16:4916–4923. https://doi.org/10.1093/emboj/16.16.4916
Kim HK, Cho EJ, Jo S, Sung BR, Lee S, Yun SH (2012) A split luciferase complementation assay for studying in vivo protein–protein interactions in filamentous ascomycetes. Curr Genet 58:179–189. https://doi.org/10.1007/s00294-012-0375-5
Kjaerbolling I, Vesth TC, Frisvad JC, Nybo JL, Theobald S, Kuo A, Bowyer P, Matsuda Y, Mondo S, Lyhne EK, Kogle ME, Clum A, Lipzen A, Salamov A, Ngan CY, Daum C, Chiniquy J, Barry K, LaButti K, Haridas S, Simmons BA, Magnuson JK, Mortensen UH, Larsen TO, Grigoriev IV, Baker SE, Andersen MR (2018) Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc Natl Acad Sci USA 115:E753–E761. https://doi.org/10.1073/pnas.1715954115
Krijgsheld P, Nitsche BM, Post H, Levin AM, Muller WH, Heck AJ, Ram AF, Altelaar AF, Wosten HA (2013) Deletion of flbA results in increased secretome complexity and reduced secretion heterogeneity in colonies of Aspergillus niger. J Proteome Res 12:1808–1819. https://doi.org/10.1021/pr301154w
Lee BN, Adams TH (1994) Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol Microbiol 14:323–334. https://doi.org/10.1111/j.1365-2958.1994.tb01293.x
Lengeler KB, Davidson RC, D’Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64:746–785. https://doi.org/10.1128/mmbr.64.4.746-785.2000
Leslie JF, Summerell BA (2006) The Fusarium laboratory manual. Blackwell Publishing Professional, Ames
Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, Naqvi NI (2007) Rgs1 regulates multiple G alpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J 26:690–700. https://doi.org/10.1038/sj.emboj.7601536
Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, Houterman PM, Kang S, Shim WB, Woloshuk C, Xie X, Xu JR, Antoniw J, Baker SE, Bluhm BH, Breakspear A, Brown DW, Butchko RA, Chapman S, Coulson R, Coutinho PM, Danchin EG, Diener A, Gale LR, Gardiner DM, Goff S, Hammond-Kosack KE, Hilburn K, Hua-Van A, Jonkers W, Kazan K, Kodira CD, Koehrsen M, Kumar L, Lee YH, Li L, Manners JM, Miranda-Saavedra D, Mukherjee M, Park G, Park J, Park SY, Proctor RH, Regev A, Ruiz-Roldan MC, Sain D, Sakthikumar S, Sykes S, Schwartz DC, Turgeon BG, Wapinski I, Yoder O, Young S, Zeng Q, Zhou S, Galagan J, Cuomo CA, Kistler HC, Rep M (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367–373. https://doi.org/10.1038/nature08850
Mukherjee M, Kim JE, Park YS, Kolomiets MV, Shim WB (2011) Regulators of G-protein signalling in Fusarium verticillioides mediate differential host–pathogen responses on nonviable versus viable maize kernels. Mol Plant Pathol 12:479–491. https://doi.org/10.1111/j.1364-3703.2010.00686.x
Park AR, Cho AR, Seo JA, Min K, Son H, Lee J, Choi GJ, Kim JC, Lee YW (2012) Functional analyses of regulators of G protein signaling in Gibberella zeae. Fungal Genet Biol 49:511–520. https://doi.org/10.1016/j.fgb.2012.05.006
Proctor RH, Desjardins AE, Plattner RD, Hohn TM (1999) A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi slating population A. Fungal Genet Biol 27:100–112. https://doi.org/10.1006/fgbi.1999.1141
Proctor RH, Van Hove F, Susca A, Stea G, Busman M, van der Lee T, Waalwijk C, Moretti A, Ward TJ (2013) Birth, death and horizontal transfer of the fumonisin biosynthetic gene cluster during the evolutionary diversification of Fusarium. Mol Microbiol 90:290–306. https://doi.org/10.1111/mmi.12362
Ramanujam R, Xu Y, Hao L, Naqvi NI (2012) Structure-function analysis of Rgs1 in Magnaporthe oryzae: role of DEP domains in subcellular targeting. PLoS ONE 7:e41084. https://doi.org/10.1371/journal.pone.0041084
Ramanujam R, Calvert ME, Selvaraj P, Naqvi NI (2013) The late endosomal HOPS complex anchors active G-protein signaling essential for pathogenesis in Magnaporthe oryzae. PLoS Pathog 9:e1003527. https://doi.org/10.1371/journal.ppat.1003527
Sagaram US, Shim WB (2007) Fusarium verticillioides GBB1, a gene encoding heterotrimeric G protein beta subunit, is associated with fumonisin B biosynthesis and hyphal development but not with fungal virulence. Mol Plant Pathol 8:375–384. https://doi.org/10.1111/j.1364-3703.2007.00398.x
Shim WB, Woloshuk CP (1999) Nitrogen repression of fumonisin B1 biosynthesis in Gibberella fujikuroi. FEMS Microbiol Lett 177:109–116. https://doi.org/10.1111/j.1574-6968.1999.tb13720.x
Wang Y, Geng Z, Jiang D, Long F, Zhao Y, Su H, Zhang KQ, Yang J (2013) Characterizations and functions of regulator of G protein signaling (RGS) in fungi. Appl Microbiol Biotechnol 97:7977–7987. https://doi.org/10.1007/s00253-013-5133-1
Woloshuk CP, Shim WB (2013) Aflatoxins, fumonisins, and trichothecenes: a convergence of knowledge. FEMS Microbiol Rev 37:94–109. https://doi.org/10.1111/1574-6976.12009
Xue C, Hsueh Y-P, Heitman J (2008) Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev 32:1010–1032. https://doi.org/10.1111/j.1574-6976.2008.00131.x
Yan H, Shim WB (2020) Characterization of non-canonical G beta-like protein FvGbb2 and its relationship with heterotrimeric G proteins in Fusarium verticillioides. Environ Microbiol 22:615–628. https://doi.org/10.1111/1462-2920.14875
Yan H, Huang J, Zhang H, Shim WB (2019) A Rab GTPase protein FvSec4 is necessary for fumonisin B1 biosynthesis and virulence in Fusarium verticillioides. Curr Genet 66:205–216. https://doi.org/10.1007/s00294-019-01013-6
Yu JH (2010) Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiol 38:229–237. https://doi.org/10.4489/MYCO.2010.38.4.229
Yu JH, Keller N (2005) Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol 43:437–458. https://doi.org/10.1146/annurev.phyto.43.040204.140214
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981. https://doi.org/10.1016/j.fgb.2004.08.001
Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, Chen Y, Wang J, Qi Z, Wang Z, Zheng X, Wang P, Zhang Z (2011a) Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog 7:e1002450. https://doi.org/10.1371/journal.ppat.1002450
Zhang Y, Choi YE, Zou X, Xu JR (2011b) The FvMK1 mitogen-activated protein kinase gene regulates conidiation, pathogenesis, and fumonisin production in Fusarium verticillioides. Fungal Genet Biol 48:71–79. https://doi.org/10.1016/j.fgb.2010.09.004
Zhang H, Mukherjee M, Kim JE, Yu W, Shim WB (2018) Fsr1, a striatin homologue, forms an endomembrane-associated complex that regulates virulence in the maize pathogen Fusarium verticillioides. Mol Plant Pathol 19:812–826. https://doi.org/10.1111/mpp.12562
Acknowledgements
This research was supported in part by the Agriculture and Food Research Initiative Competitive Grants Program Grant (2013-68004-20359) from the USDA National Institute of Food and Agriculture. We also would like to acknowledge China Scholarship Council support (201906850058) for Z. Zhou. The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
294_2020_1140_MOESM1_ESM.pdf
Supplementary file 1: Fig. S1. Sequence alignment FlbA ortholog proteins in select fungal species. We aligned protein sequences of Fusarium verticillioides FvFlbA1, F. verticillioides FvFlbA2, Saccharomyces cerevisiae ScSst2, A. nidulans AnFlbA, Neurospora crassa NcFlbA, and Magnaporthe oryzae MoRgs1. Identical and similar sequences were displayed in the box. Fig. S2. Split marker approach employed in FLBA generating gene deletion mutants in F. verticillioides. (A) ΔFvflbA1 mutant was generated by replacing FLBA1 gene with a hygromycin B phosphotransferase gene (HPH). (B) FvFLBA2 gene was replaced by a hygromycin gene to generate a ΔFvflbA2 mutant. (C) ΔFvflbA2/A1FvFLBA1 was generated by replacing FLBA1 gene with a geneticin gene (GEN) in ΔFvflbA2 background. (D) Mutants ΔFvflbA1, ΔFvflbA1-Com, ΔFvflbA2/A1 were subjected to qPCR using FvFLBA1 gene primer. Relative expression was normalized to F. verticillioides β-tubulin gene (FVEG_04081). (E) FvFLBA2 gene transcription level was examined in WT, ΔFvflbA2, ΔFvflbA2-Com, ΔFvflbA2/A1 strains. Fig. S3. Relative FB1 expression in wild-type and FvflbA mutants. FB1 was extracted from supernatant (2ml) of 7-day incubation at 150 RPM in myro liquid culture. In details, YEPD mycelial samples (3 dpi) were harvested through Miracloth (EMD Millipore). Subsequently, mycelia (0.3g) were inoculated into 100 ml myro liquid medium at room temperature. (PDF 240 KB)
Rights and permissions
About this article
Cite this article
Yan, H., Zhou, Z. & Shim, W.B. Two regulators of G-protein signaling (RGS) proteins FlbA1 and FlbA2 differentially regulate fumonisin B1 biosynthesis in Fusarium verticillioides. Curr Genet 67, 305–315 (2021). https://doi.org/10.1007/s00294-020-01140-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-020-01140-5