Abstract
Candida auris is a newly emerged pathogenic microbe, having been identified as a medically relevant fungus as recently as 2009. It is one of the most drug-resistant yeast species known to date and its emergence and population structure are unusual. Because of its recent emergence, we are largely ignorant about fundamental aspects of its general biology, life cycle, and population dynamics. Here, we report the karyotype variability of 26 C. auris strains representing the four main clades. We demonstrate that all strains are haploid and have a highly plastic karyotype containing five to seven chromosomes, which can undergo marked alterations within a short time frame when the fungus is put under genotoxic, heat, or osmotic stress. No simple correlation was found between karyotype pattern, drug resistance, and clade affiliation indicating that karyotype heterogeneity is rapidly evolving. As with other Candida species, these marked karyotype differences between isolates are likely to have an important impact on pathogenic traits of C. auris.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A current, major concern in medical mycology is the emergence of the multidrug-resistant pathogen Candida auris. This species was named according to its first identification as an isolate from the ear canal of a Japanese patient in 2009 (Satoh et al. 2009). Since then, it has rapidly become a major healthcare threat with hospital outbreaks occurring worldwide (Chowdhary et al. 2016; Jeffery-Smith et al. 2018). Most C. auris isolates show high levels of resistance to antifungal drugs, including azoles, echinocandins, 5-flucytosine, and polyenes (amphotericin B) (Chakrabarti et al. 2014; Lockhart et al. 2017). C. auris is also difficult to eradicate from hospital intensive care wards and as a skin colonizer it can apparently be transmitted from patient to patient (Jeffery-Smith et al. 2018).
Whole-genome sequencing (WGS) of C. auris isolates has indicated that there are at least four distinct geographical clades of this species; East Asia (Japan, Korea), South Asia (India, Pakistan), South Africa, and South America (Venezuela) (Lockhart et al. 2017). Clades differ by tens of thousands of single-nucleotide polymorphisms (SNPs) from each other; however, within each clade, isolates are almost indistinguishable from each other on a DNA sequence level (Chakrabarti et al. 2014; Lockhart et al. 2017; Rhodes et al. 2018). This suggests that the C. auris population structure is characterized by distinct and highly variable clades that are distributed worldwide and almost non-variable clonal expansions of a single genotype within individual outbreaks (Jeffery-Smith et al. 2018). The origin(s) of the strong variability between and the minor variability within clades are currently unknown.
Polyploidy, aneuploidy, and gross chromosome rearrangements have been recognized as drivers of genetic diversity in pathogenic and non-pathogenic fungi for some time (Zolan 1995; Fierro and Martín 1999; Bennett et al. 2014; Wertheimer et al. 2016; Monerawela and Bond 2017; Harari et al. 2018a). In pathogenic yeasts, such as C. albicans, mechanisms for ploidy shifts and chromosome rearrangements have been described, and their importance for adaptation to environmental stresses and host niches, as well as for developing resistance to antifungal drugs has been identified (Selmecki et al. 2006, 2009; Wertheimer et al. 2016).
Here, we characterize a set of 26 clinical isolates of the newly emerging human pathogenic fungus C. auris to understand whether its genome has undergone structural alterations potentially underlying adaptation events. This strain collection covers all four geographical clades, different levels of drug resistance, and various sources of isolation (Table S1). All isolates were shown to be haploid, and we observed substantial karyotypical variability between C. auris strains, even between isolates belonging to the same clade. Importantly, genetic diversity on a DNA sequence level within a clade had been reported to be minimal (Chakrabarti et al. 2014; Lockhart et al. 2017; Rhodes et al. 2018). We also tested whether under heat, osmotic, or DNA replication stress karyotype changes are induced in C. auris, similar to other fungi (Todd et al. 2017). The frequency of changes is higher in stress conditions, but minor alterations could also appear when C. auris was grown in standard laboratory conditions. We observed that in some cases these changes are associated with fitness benefits. However, other karyotype modifications seem to be stochastic and would not confer an advantage, as previously reported for other fungi (Rustchenko et al. 1997; Janbon et al. 1999).
Materials and methods
Yeast strains and culture
Candida auris and other yeast strains used in this study are listed in Table S1. Candida albicans SC5314, and Saccharomyces cerevisiae BY4741 and BY4743 were used as control organisms. Yeast cells were grown at 30 °C on YPD plates (1% yeast extract, 2% mycological peptone, 2% glucose, 2% agar; Oxoid, Basingstoke, UK) or shaking at 200 rpm in YPD broth (same as plates, but without agar).
Flow cytometry
Processing yeast samples for flow cytometry was performed largely as previously described (Fortuna et al. 2001). Briefly, stationary-phase yeast cells were inoculated into fresh YPD broth and incubated while shaking (200 rpm) at 30 °C for 3 h. Cells were harvested by centrifugation (1000 × g, 2 min), re-suspended at a concentration of 1 × 107 cells/ml in ice-cold demineralized water, and fixed overnight by adding 100% ethanol to a final concentration of 70% ethanol. Cells were then harvested by centrifugation (1000 × g, 2 min) and re-suspended in 50 mM sodium citrate (pH 7.5). After RNase A (250 μg per 1 × 107 cells) and proteinase K (1000 μg per 1 × 107 cells) treatment, cells were transferred to 12 × 75 mm round-bottom tubes. After adding Triton-X 100 (Sigma-Aldrich) to a final concentration of 0.25%, and SYBR Green I (1:500; Sigma-Aldrich, St. Louis, MI, USA) as a DNA stain, samples were incubated at 4 °C overnight. Before flow cytometry, samples were sonicated (three pulses at 30–60 W for 1–2 s; Sonicator Ultrasonic Processor S-4000, Misonix, Farmingdale, NY, USA). Flow cytometry was performed on a BD LSR II flow cytometer (BD Biosciences, San Jose, CA, USA) using an excitation wavelength of 488 nm, SYBR Green I fluorescence was detected with a 530/30 band pass filter. 10,0000 events, gated for singlets, were recorded for every sample. Data were analyzed using FlowJo 10.2 software (FlowJo LLC, Ashland, OR, USA).
Pulsed-field gel electrophoresis (PFGE)
Chromosomal DNA of C. auris strains was embedded in agarose plugs using the CHEF Genomic DNA Plug Kit (Bio-Rad Laboratories Ltd., Hercules, CA, USA) following the instructions of the manufacturer. For some strains, the cell wall digestion reaction was supplemented with Lallzyme MMX (end concentration 100 mg/ml; Lallemand Inc., Quebec, Canada). Pulsed-field gel electrophoresis (PFGE) was performed on a CHEF Mapper XA System (Bio-Rad). As a standard programme C. auris DNA was run for 48 h at 14 °C in 1 × TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA; pH 8.0) and 0.8% Megabase agarose (Bio-Rad) at 3.0 V/cm applied at a 106° angle and a switch time of 500 s; every strain and isolate were run at least twice under these conditions (images are available at https://dx.doi.org/10.6084/m9.figshare.7881167). To get a better separation of smaller chromosomes, DNA from selected C. auris strains was run for 48 h at 14 °C in 1 × TAE and 0.8% Pulsed-Field Certified agarose (Bio-Rad) at 4.0 V/cm applied at a 120° angle, initial and final switch times of 120 s and 240 s using linear ramping. Gels were stained with SYBR Green I (Sigma-Aldrich) diluted 1:10,000 in 1 × TAE for at least 1 h and documented by photography under UV illumination on a Gel Doc EQ system controlled by Quantity One software (version 4.6.6) (Bio-Rad).
Southern blot analysis
Chromosome-sized DNA bands from PFGE gels were transferred to Zeta-Probe GT membranes (Bio-Rad) by alkaline Southern blotting following previously described principles (Sambrook and Russell 2000). Gels were soaked in depurinating solution (0.25 M HCl) for ~ 25 min and, after that, in denaturing solution (1.5 M NaCl, 0.5 M NaOH) for ~ 30 min. Using capillary transfer in denaturing solution for 24 h chromosomal DNA was immobilized on the membranes. After transfer, membranes were steeped in neutralization buffer (0.5 M Tris, pH 7.0) for 5 min, washed briefly in 2 × SSC (saline–sodium citrate; 300 mM NaCl, 30 mM Na3C6H5O7, pH 7.0) and dried at room temperature.
A fragment of 836 bp was amplified by polymerase chain reaction (PCR) from the 25S rRNA region of C. auris strain UACa11 genomic DNA using oligonucleotides oUA367 (5′-GGCAAAACAAAGGCCGCGC-3′) and oUA368 (5′-AGTAGCTGGTTCCTGCCGAAG-3′). This fragment was used as template for a labeling PCR incorporating digoxigenin-11-dUTP with nested primers oUA371 (5′-CCAATTCCAGGGTCACAGGCT-3′) and oUA372 (5′-CCTCAGGATAGCAGAAGCTCGT-3′) to give an rDNA probe of 759 bp (DIG DNA Labeling Mix; Roche Molecular Systems Inc., Pleasanton, CA, USA). All PCR reactions were carried out using GoTaq® G2 Flexi DNA Polymerase (Promega Corp., Madison, WI, USA). Oligonucleotides were supplied by Sigma-Aldrich Co. (St. Louis, MO, USA).
Membranes were hybridized with the digoxigenin-11-dUTP labeled rDNA probe using DIG DNA Labeling Kit (Roche Molecular Systems Inc.), and then incubated with α-digoxigenin antibody (Roche Molecular Systems Inc.) conjugated to alkaline phosphatase. Alkaline phosphatase bound to digoxigenin-11-dUTP labeled bands was then detected on a FUSION SL Chemiluminescence Imaging System (Vilber Lourmat, Marne-la-Vallée, France) using CPD-Star chemi-luminescent substrate (Roche Molecular Systems Inc.).
Microevolution assay
A microevolution assay was carried out using selected strains from each clade (UACa11, UACa18, UACa20, and UACa22) to test whether karyotype variation can be induced by particular growth conditions: strains were separately passaged five times through YPD broth at 30 °C (control), YPD broth at 42 °C (heat stress), synthetic-defined liquid medium containing 2% sorbose (SSD) (6.7 g/l yeast nitrogen base with amino acids, Sigma-Aldrich) at 30 °C (osmotic stress), and YPD broth containing 100 mM hydroxyurea (HU; Formedium, Norfolk UK) at 30 °C (DNA replication stress). All passages were performed following the same experimental strategy (Fig. S1). Parental strains, from glycerol stock, were grown on YPD plates overnight at 30 °C; ~ 107 cells from this plate were used to initiate the different passages (Fig. S1). A passage consisted of (1) growing cells in liquid culture overnight under treatment conditions, (2) plating 100–200 cells from that liquid culture on appropriate solid medium until single colonies were visible (2–3 days, except on 2% sorbose where incubation took 1 week), (3) suspending the five largest single colonies in 1 ml of sterile water, determining cell concentration, and making four 1:10 serial dilutions, from 102 to 105 cells, to test resulting isolates in spot assays under the same conditions, (4) selecting the three fastest-growing isolates for long-term storage (40% glycerol at − 70 °C), and (5) inoculating the fastest-growing isolate at ~ 107 cells in a fresh overnight liquid culture starting the next passage (Fig. S1).
Isolates from the first and the fifth passage of each strain and condition were subjected to PFGE analysis (see above), and spot assays under the same conditions used for each passage (except for DNA replication stress where isolates were also tested on plates containing a higher concentration of 200 mM HU); the parental strain was always included for comparison. Isolates from the fifth passage in sorbose were also tested on YPD plates containing 4 and 16 μg/ml caspofungin (CSP) at 30 °C. For the DNA preparation for PFGE analysis and the spot assays, isolates and parental strains were revived from long-term storage on YPD agar (overnight at 30 °C) and then grown in YPD broth overnight at 30 °C. Appropriate plates were grown for 1–6 days depending on conditions and temperature used for each passage.
Cell concentrations were determined by measuring optical density of the culture at a wavelength of 600 nm (OD600) on an Ultraspec 2000 (Pharmacia Biotech, Sweden) spectrometer. Previous calibration defined a C. auris culture of OD600 = 1 to contain 3 × 107 cells/ml.
Results
Candida auris is a haploid fungus
Polyploidy and complex aneuploidy play a major role in the capability of fungal pathogens to adapt to various stresses and to the changing condition within host niches (Bennett et al. 2014; Wertheimer et al. 2016). These overall genome shifts in chromosome number have been characterized as drivers of increased genetic diversity in Cryptococcus neoformans, Candida albicans, and also in Candida lusitaniae—a close relative of C. auris (Forche et al. 2008; Reedy et al. 2009; Ni et al. 2013; Hirakawa et al. 2017).
Therefore, we were interested in determining the ploidy of clinical C. auris isolates to understand whether chromosome number variations and whole-genome duplication potentially are adaptive strategies employed by C. auris. In total 25 C. auris strains covering all four geographical clades, comprising antifungal-sensitive, -resistant, and -multiresistant isolates from various sources of infection, as well as strains from a single outbreak (Royal Brompton hospital, London, UK) (Table S1) were tested by flow cytometry (Chakrabarti et al. 2014; Sharma et al. 2015; Schelenz et al. 2016; Lockhart et al. 2017). Haploid and diploid Saccharomyces cerevisiae strains and the diploid C. albicans laboratory strain SC5314 were used as references. Estimates from whole-genome sequencing suggested that C. auris has a similar genome size as S. cerevisiae at approx. 12 Mbp (Goffeau et al. 1996; Lockhart et al. 2017). As expected, C. albicans SC5314 had a similar cell cycle profile as a diploid S. cerevisiae strain (Fig. 1). In contrast, the genome size of all 25 C. auris strains was found to be consistent with containing a haploid chromosome complement (Fig. 1), although the resolution of flow cytometry probably does not allow us to unequivocally exclude the occasional disomy of one of the smaller chromosomes. However, in none of the 25 strains, disomies were apparent in the karyotype analysis (see below).
Cell cycle profiles of clinical Candida auris isolates. Histogram showing cell cycle profiles obtained by flow cytometry after staining DNA with SYBR green of 25 C. auris strains (Table S1). Diploid Candida albicans (Calb) strain SC5314, haploid (n, BY4741) and diploid (2n, BY4743) strains of Saccharomyces cerevisiae (Sc) were included as controls. C. auris has the same cell cycle profile as haploid S. cerevisiae. C. auris strains are grouped according to their taxonomical position within the four geographical clades: E. Asia (UACa7 & UACa18), S. Asia-India (UACa1-5, UACa8, UACa9, UACa11, UACa26-27), S. Asia-Pakistan (UACa19, UACa24-25), strains from the Royal Brompton hospital outbreak (UACa12-15), S. Africa (UACa6, UACa10, UACa20-21), and S. America (UACa22-23). Approximate position of haploid G1 (1C), haploid G2 and diploid G1 (2C), and diploid G2 (4C) peaks are indicated at the top
This result indicates, that major ploidy changes do not appear to be a mechanism C. auris employs to adapt to the environmental challenges tested here. If it does, this must be only temporarily with diploids, polyploids, or aneuploids returning quickly to a haploid stage after these challenges are removed.
Candida auris clinical isolates have a plastic karyotype
Since all the C. auris isolates tested turned out to be haploid (or near haploid), we wondered whether its genetic diversity might be generated by gross chromosome rearrangements. To test this we utilized PFGE to separate C. auris chromosomes (Fig. 2). We characterized the same 25 strains covering all four geographical clades and the single hospital outbreak as above, as well as an additional East Asian isolate, UACa83 (the type-strain of C. auris, CBS10913T) (Table S1) (Satoh et al. 2009; Chakrabarti et al. 2014; Sharma et al. 2015; Schelenz et al. 2016; Lockhart et al. 2017). 11 of these strains have previously been whole-genome sequenced (Sharma et al. 2015; Lockhart et al. 2017), and two of these (UACa20, and UACa24) had their genomes assembled into seven chromosome-size contigs (Muñoz et al. 2018). C. auris isolates show chromosome numbers from 5 to 7, ranging from ~ 0.7 Mbp to ~ 3.25 Mbp in size (Figs. 2, 3); chromosomes will be referred to by their size. Additionally, we probed the karyotypes for the location of the repetitive rRNA gene clusters (rDNA) by Southern blotting (Figs. 2, 3). It should be noted that chromosomal bands of similar size are difficult to separate using the PFGE system; however, we have considered the presence of two chromosomes when the band intensity is clearly higher, e.g., chromosomal band around 1 Mbp in UACa20 likely or chromosomal band around 1.35 Mbp in UACa18 possibly each contain two chromosomes.
Representative PFGE karyotypes of clinical Candida auris isolates. PFGE karyotypes (left panels, inverted image) and associated Southern blots using a rDNA probe to detect rRNA gene clusters (right panels) of the indicated strains representing examples of E. Asian (UACa7, UACa18, UACa83), S. Asian (UACa4, UACa5, UACa9, UACa11, UACa25, UACa26, UACa27), S. African (UACa10, UACa20), and S. American (UACa22) clades, as well as one isolate from the outbreak at the Royal Brompton Hospital, London, UK (UACa13) (Table S1). a Gel run at standard conditions (0.8% Megabase agarose, 1 × TAE, 48 h, 14 °C, 3.0 V/cm, 106°, switch time 500 s), Hansenula wingei (Hw) CHEF DNA size marker (Bio-Rad) serving as standard (size of chromosomal bands in Mbp indicated on the left). Single bands considered as two chromosomes are indicated by red asterisks (*). b Gel run at conditions to resolve smaller chromosomes (0.8% Pulsed Field Certified agarose, 1 × TAE, 48 h, 14 °C, 4.0 V/cm, 120°, switch times: linear ramping 120–240 s), Saccharomyces cerevisiae (Sc) CHEF DNA size marker (Bio-Rad) serving as standard (size of some chromosomal bands in Mbp indicated on the left)
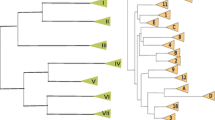
Schematic of karyotypes of 26 clinical Candida auris isolates. Strains are grouped according to their taxonomical position within the four geographical clades. Chromosomes are represented as differently coloured rectangles to indicate affiliation to a particular clade: E. Asia (yellow), S. Asia (India, red; Pakistan, light red), S. Africa (green), and S. America (blue); the four strains isolated from the outbreak at the Royal Brompton Hospital, London (Brompton) are indicated as a separated group (purple). Black rectangles indicate the chromosomes of Hansenula wingei (Hw). Chromosome sizes were measured using the Hansenula wingei (Hw) CHEF DNA size marker (Bio-Rad) and strain UACa11 as standards, both of which were run alongside the samples in each gel. Chromosomes harboring rRNA gene clusters are indicated by a black circle (●). See https://dx.doi.org/10.6084/m9.figshare.7881167 for gel images
The E. Asian strains of this study apparently have five to seven chromosomes one of which carries rRNA gene repeats. Intriguingly, UACa18 (B11220 obtained from the CDC, Atlanta, GA, USA) and UACa83 (CBS10913T obtained from the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands) originate from the same type material (Satoh et al. 2009), but do show slight karyotypical differences in the smaller chromosomes (Fig. 3). The S. African and S. American isolates we studied all had seven chromosomes and with the exceptions of UACa6 (S. Africa) and UACa22 (S. America) only one of the chromosomes was rDNA-bearing (Fig. 3). In contrast, chromosome numbers in S. Asian isolates ranged from six to seven, and, except for UACa9, all S. Asian strains had at least two chromosomes carrying rRNA gene repeats (Fig. 3). The range of chromosome numbers and of chromosome size distributions in S. Asian isolates may reflect the comparatively large intra-clade genetic diversity of this strain cluster (Lockhart et al. 2017), but we cannot exclude this to be an issue of having only a small sample set available for non-S. Asian isolates. The four strains from the Royal Brompton hospital outbreak (UACa12–UACa15) had similar karyotypes to each other with seven detectable chromosomal bands. In these strains the two chromosomes bearing rDNA repeats displayed subtle size differences between these particular set of strains (Fig. 3). Interestingly, the four strains from the Royal Brompton hospital outbreak appeared similar to S. Asian strains, which they are also most closely related to according to whole-genome sequence analysis (Rhodes et al. 2018).
Adding chromosome-size estimates from PFGE points towards a range of genome sizes between ~ 10 Mbp and ~ 13 Mbp which is a reasonable fit to the 12.5 Mbp suggested by whole-genome sequencing (Lockhart et al. 2017), and conforms with our flow cytometry results (Fig. 1). Complete assembly of whole-genome sequences into chromosome-sized contigs to create physical maps of C. auris genomes will allow full appreciation of the genome structure of this fungus. Indeed, a recent study reported seven contigs for two C. auris isolates, in our PFGE analysis the corresponding strains UACa20 (B11221) and UACa24 (B8441) also display seven chromosomal bands similar in size to the reported contigs (Muñoz et al. 2018).
Electrophoretic karyotyping revealed that C. auris isolates differed considerably in chromosome numbers and sizes, both within and between geographical clades. This plasticity was somewhat unexpected considering the genetic uniformity of C. auris on a DNA sequence level within geographical clades and within hospital outbreaks, and indicates that gross chromosome rearrangements might be a mechanism C. auris employs to generate genetic diversity during adaptation to environmental challenges (Sharma et al. 2016; Lockhart et al. 2017; Rhodes et al. 2018).
C. auris undergoes karyotype rearrangements in stress conditions
To get insight about C. auris fitness and its relation to the karyotype variation observed in different clinical isolates, four strains (UACa11, UACa18, UACa20, and UACa22), one from each clade (Table S1), were selected to undergo a microevolution experiment. Strains were grown under four different conditions through five passages each (see “Materials and Methods”, Fig. S1): standard YPD broth at 30 °C (control); heat stress at 42 °C; osmotic stress using 2% sorbose, which mimics the effect of echinocandin-type antifungals; and DNA replication stress using hydroxyurea (HU), an inhibitor of the enzyme ribonucleotide reductase which depletes nucleotide pools (Koç et al. 2004; Yang et al. 2013). These conditions have been described previously as factors inducing genome instability in fungi. Introduction of heat stress has been shown to induce ploidy variation in C. albicans and other fungal species (Anderson et al. 2015). In C. albicans, treatment with 2% sorbose is a classic example for inducing changes in the karyotype, which offer a phenotypic advantage in this stress (Rustchenko et al. 1994; Janbon et al. 1999; Kabir et al. 2005). Replication stress, for example by treatment with HU, has been associated with genome instability in a wide range of organisms, including yeast and human cells (Arlt et al. 2011; Maxwell 2016).
In our microevolution study, karyotype modifications including appearance, disappearance, or size changes of different chromosomal bands were observed in all tested strains. However, the frequency and type of modification were different depending on the strain and condition used (Fig. 4).
Karyotype variation during microevolution of Candida auris isolates. Schematic representation of karyotypes (top panels) and spot assays (bottom panels) of four C. auris isolates covering all four established clades (a) UACa11 (S. Asia), (b) UACa18 (E. Asia), (c) UACa20 (S. Africa), and (d) UACa22 (S. America)—obtained after the first and fifth passages in four different growth conditions: YPD at 30 °C; YPD at 42 °C, heat stress (HE); YPD containing 100 mM hydroxyurea at 30 °C (HU); and 2% sorbose in synthetic defined medium at 30 °C (sorbose). Chromosomes are represented as coloured rectangles for different strains or black rectangle for Hansenula wingei (Hw) CHEF DNA size marker (Bio-Rad) used as a chromosome size standard (numbers represent size in Mbp). Chromosomal bands that seemingly disappear in comparison to parental strains are represented as empty rectangles with dotted lines. Darker rectangles represent new chromosomal bands appearing in comparison to parental strains. In strain UACa20 under heat stress the karyotype changes observed indicate massive chromosome rearrangements. Chromosomes harboring rRNA gene clusters are indicated by a black circle (●) only in parental strains. Spot assays show the parental strain (P) and derived isolates for comparison, grown in the same conditions used for the respective passages, except for HU for which a higher concentration (200 mM) than during the passages (100 mM) is used. Serial dilutions contain 105, 104, 103, and 102 cells. Isolates were grown for 1 day in heat stress (42 °C), 3 days in HU (due to slow growth, UACa18 and its derivatives were grown for 6 days), or 3 days in 2% sorbose. See https://dx.doi.org/10.6084/m9.figshare.7881167 for gel images
The S. Asian strain UACa11 has shown chromosome modifications under heat and DNA replication (HU) stress conditions, always related to chromosomes carrying rDNA repeats (Fig. 4a). In HU, the 1.35 Mbp chromosomal band seemingly disappears after only one passage. In heat stress, the 1.35 Mbp and 1.6 Mbp chromosomal bands disappeared, and two additional ones appeared (around 1.1 Mbp and 1.7 Mbp), most likely due to loss and gain of DNA from the corresponding original chromosomes.
The E. Asian isolate UACa18 showed a tendency to lose the 0.95 Mbp chromosomal band in all stress conditions, but not when growing in YPD at 30 °C (control), most likely changing in size to a slightly smaller band of around 0.9 Mbp (Fig. 4b). When treated with sorbose, a new chromosome of around 1.3 Mbp appeared after five passages in UACa18, likely because of the reduction in size of one of the two 1.35 Mbp chromosomes.
Looking at the S. African strain UACa20, only minor changes were observed after five passages in sorbose and HU (Fig. 4c). Specifically, one alteration was observed in sorbose where one of the isolates seemed to gain DNA in the 1.65 Mbp chromosome, in HU where the 1.05 Mbp chromosome decreased in size in one isolate, and two other chromosomes (1.6 and 1.65 Mbp) seemingly reduced size in another isolate. However, under heat stress conditions drastic changes were observed, the number of chromosomes reduced from seven to six, four, or three bands in three different isolates. Importantly, flow cytometry demonstrated that these substantial karyotype changes did not cause any conspicuous alterations of the ploidy state (Fig. S2).
Finally, the S. American strain UACa22 was the most plastic, showing karyotype changes even when grown in control conditions (YPD, 30 °C), in this case chromosomes carrying the rDNA tend to change size, losing or gaining DNA (Fig. 4d). In almost all conditions the 1.2 Mbp chromosome tends to reduce in size. Besides, a second chromosomal band (1.6 Mb) was seemingly lost in the presence of HU after five passages.
Despite the karyotype alterations observed, a moderate improvement in growth was detected only in a few cases (Fig. 4): isolates from UACa18 microevolved in 2% sorbose from the first passage, two isolates from UACa18 after one passage in 100 mM HU; and isolates from UACa22 after five passages of 100 mM HU when tested at a higher concentration of 200 mM HU. These improvements in growth could be correlated with changes in the karyotype in some cases (see above). However, this clearly is not probable for all the cases, e.g., isolates of strain UACa20 that grow better in sorbose, but did not show any difference in karyotype in comparison to the parental strain, and modifications in karyotype that did not obviously offer any improved fitness in the condition tested. Interestingly, the massive karyotype variation observed in microevolved UACa20 isolates obtained under heat stress did not show an apparent difference growing at 42 °C compared to the parental strain. Isolates from the strain UACa11 did not show improvement in growth under any conditions. Because osmotic stress in 2% sorbose mimics the effect of echinocandins, we tested whether growth of isolates obtained from the fifth passage in 2% sorbose displayed improved growth in the presence of caspofungin (CSP) (Fig. S3). Isolates derived from the CSP-sensitive UACa18 strain had undergone karyotype alterations and showed a clear improvement in growth both on 2% sorbose and in the presence of CSP (Figs. 4b, S3). However, this was different for the other microevolved isolates: derivatives of UACa11 showed slow growth already on 4 μg/ml CSP in comparison to the parental strain (Fig. S3), without any distinguishable growth defect on 2% sorbose (Fig. 4a). Isolates from strains UACa20 and UACa22 were highly resistant to CSP similar to the parental strains, and original and derived isolates showed little differences in growth on 2% sorbose (Figs. 4c, d, S3).
Discussion
Here, we show that the genome of C. auris undergoes substantial karyotypic reorganization under stress conditions (Fig. 4), similar to other fungi (Ormerod and Fraser 2013; Ahmad et al. 2014; Wertheimer et al. 2016). Karyotype variation between different strains of the same species is common in diploid C. albicans (Chu et al. 1993; Magee and Magee 1997) and haploid Schizosaccharomyces pombe (Brown et al. 2011; Jeffares et al. 2017), and it has been proposed as a quick solution for adaptation to environmental changes. This variation of genome organization can take on different expressions, e.g., ploidy variation, chromosome copy number variation, or gain and loss of supernumerary chromosomes (Covert 1998; Tang and Amon 2013; Bennett et al. 2014; Todd et al. 2017; Zhang and Ma 2017; Harari et al. 2018b). High levels of genetic diversity can be introduced into a population by changes in ploidy. In yeast most of these changes produce a mixture of aneuploid populations offering a rapid solution for stress adaptation, this has been suggested to be the norm in fungi (Bennett et al. 2014). These changes in genome structure might be introduced by various mechanisms usually related to chromosome segregation mistakes during mitosis, meiosis, or during parasexual reproduction. Once ploidy changes arise in S. cerevisiae, rates of chromosome loss, genetic mutation, and microsatellite instability increase, usually leading to proliferative disadvantages (Torres et al. 2007, 2010; Forche et al. 2008; Sheltzer and Et 2011). Having said that, aneuploid and polyploid isolates of S. cerevisiae exposed to a range of stress conditions can display a growth advantage (Pavelka et al. 2010; Harari et al. 2018b); however, aneuploidy is often lost, when the stress is eliminated (Janbon et al. 1998). C. auris is haploid (Fig. 1), and so far, neither polyploid states nor sexual reproduction have been described. Therefore, we hypothesize that C. auris is likely not capable of generating genome diversity via aneuploidy or polyploidy. Further studies to elucidate the life cycle of C. auris will be necessary to shed light on this issue.
As a haploid species, the variation observed in C. auris would most likely be due to gross chromosome rearrangements, and/or possibly copy number variation (CNV) events of chromosomal sections. CNVs are a frequent reason for changes in the genome organization of haploid fungi (Zhang et al. 2013; Ahmad et al. 2014). CNVs of yet-to-be-identified genomic regions, potentially repetitive, could explain the changes in size observed in the karyotype of C. auris, e.g., the 0.95 Mbp chromosomal band in UACa18, or 1.2 Mbp chromosome in UACa22 (Fig. 4). In general, repetitive regions are known as a principal reason for CNVs and gross chromosome rearrangements in yeasts, such as transposons (Ty elements and solo LTR elements in S. cerevisiae), telomeres, or rDNA (Mieczkowski et al. 2006; Maxwell et al. 2011; Zhang et al. 2013; Kobayashi 2014; Kupiec 2014; Liu et al. 2016). Intriguingly, the appearance of CNVs increases environmental adaptation, for example increased resistance to antifungal azoles in C. albicans (Dunham et al. 2002; Selmecki et al. 2006; Measday and Stirling 2016; Hull et al. 2017). One of the best-studied repetitive elements is the highly conserved rDNA locus consisting of a large number of tandemly repeated rRNA genes interspersed with non-coding intergenic regions. In S. cerevisiae the rDNA consists of ~ 150 tandem copies of a 9.1-kb sequence, and in C. albicans the haploid genome contains a single ~ 12-kb rDNA region located on chromosome R (Keil and Roeder 1984; Jones et al. 2004). The rDNA array is highly recombinogenic undergoing fluctuations in copy number, which in S. cerevisiae is related to the loss of global chromosomal stability, especially during senescence (Kobayashi 2008; Pal et al. 2018). Furthermore, in C. albicans chromosome R has been described as more unstable than the other chromosomes within the complement, and thus more frequently displaying size changes (Iwaguchi et al. 1992; Rustchenko et al. 1993). Indeed, a set of four strains from the outbreak at the Royal Brompton hospital shows moderate size changes in the chromosomes harboring the rDNA repeats only (Fig. 3). In the first draft genome obtained for C. auris strain Ci6684, seven loci containing rRNA gene repeats were described, since this is an incomplete assembly containing 99 scaffolds, the true number of rDNA loci will likely be smaller (Chatterjee et al. 2015). Our survey of PFGE karyotype by Southern blotting using C. auris rDNA as a probe indicates that between one and four chromosomes harbor rRNA gene repeats (Fig. 3), which could be the source for some of the rearrangements we observed (Figs. 3, 4). The involvement of repetitive elements other than rDNAs, such as retrotransposon or minisatellites, in chromosome rearrangements and CNVs in C. auris, will thus be an interesting aim for future study. As example, the appearance of a novel chromosomal band (1.3 Mb) has been observed in the strain UACa18 after passaging through 2% sorbose (Fig. 4). There are several potential explanations for this observation, among the more likely ones are, (1) that the chromosomal band at 1.37 Mbp contains two chromosomes, (2) that in the microevolved UACa18-derivative the resulting population represents a mixture containing cells with the original rDNA-bearing 1.37 Mbp chromosome and cells with a considerably shorter version of this chromosome, or (3) similar to C. glabrata, that the appearance of the novel chromosome originated from segmental duplications in one of the two smaller chromosomes (Fig. 4) (Poláková et al. 2009). These novel chromosomes in C. glabrata carry duplicated genes potentially involved in yeast–host interaction and virulence (Poláková et al. 2009).
In our microevolution study, we observed the disappearance of chromosomal bands, which cannot obviously be explained by a change in size, like in strain UACa11 and UACa22 in HU (Fig. 4). In these instances, it is likely that a change in size occurred, and the new size is being masked by another chromosomal band. Due to C. auris being haploid, we can exclude a complete loss of a chromosome, as has been described in diploid C. albicans and S. cerevisiae (Selmecki et al. 2006; Wertheimer et al. 2016; Tutaj et al. 2019).
Strikingly, we observed massive chromosome rearrangements without changes in genome size in the strain UACa20 during our microevolution assay under heat stress reducing the number of chromosomes from seven down to three chromosomes in one isolate (Figs. 4, S2). This demonstrates that drastic modifications of the genome structure do not necessarily impinge on viability in C. auris, but might actually provide opportunities for general fitness adaptation. In fungi, two mechanisms have been suggested as a cause for reduction in chromosome numbers, telomere-to-telomere fusions and inactivation of one centromere, or breakage of a chromosome at a centromere and posterior fusion to telomeres of another chromosome (Gordon et al. 2011; Wendland and Walther 2014). Viable strains of S. cerevisiae with a genome consisting of only one or two chromosome have been obtained by CRISPR-Cas9-mediated engineering of end-to-end chromosome fusions and centromere deletions, though these strains display a somewhat reduced fitness (Luo et al. 2018; Shao et al. 2018). The isolates obtained from C. auris in this study with three, four, or six chromosomes, instead of seven as in the parental strains, did not show any obvious growth defects, and likely are thus fully viable (Fig. 4). Although changes in the karyotype of C. auris are not obviously faster than in any other related fungus, our results demonstrate that it is capable of undergoing and maintaining drastic alterations of its genome structure. This could be a source of adaptation to stressful conditions, and could underpin the virulence of this dangerous fungus.
References
Ahmad KM, Kokošar J, Guo X et al (2014) Genome structure and dynamics of the yeast pathogen Candida glabrata. FEMS Yeast Res 14:529–535. https://doi.org/10.1111/1567-1364.12145
Anderson CA, Roberts S, Zhang H et al (2015) Ploidy variation in multinucleate cells changes under stress. Mol Biol Cell 26:1129–1140. https://doi.org/10.1091/mbc.E14-09-1375
Arlt MF, Ozdemir AC, Birkeland SR et al (2011) Hydroxyurea induces de novo copy number variants in human cells. Proc Natl Acad Sci USA 108:17360–17365. https://doi.org/10.1073/pnas.1109272108
Bennett RJ, Forche A, Berman J (2014) Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb Perspect Med 4:a019604. https://doi.org/10.1101/cshperspect.a019604
Brown WR, Liti G, Rosa C et al (2011) A geographically diverse collection of Schizosaccharomyces pombe isolates shows limited phenotypic variation but extensive karyotypic diversity. G3 1:615–626. https://doi.org/10.1534/g3.111.001123
Chakrabarti A, Sood P, Rudramurthy SM et al (2014) Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. https://doi.org/10.1007/s00134-014-3603-2
Chatterjee S, Alampalli SV, Nageshan RK et al (2015) Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genom 16:686. https://doi.org/10.1186/s12864-015-1863-z
Chowdhary A, Voss A, Meis JF (2016) Multidrug-resistant Candida auris: “new kid on the block” in hospital-associated infections? J Hosp Infect 94:209–212. https://doi.org/10.1016/j.jhin.2016.08.004
Chu WS, Magee BB, Magee PT (1993) Construction of an Sfil macrorestriction map of the Candida albicans genome. J Bacteriol 175:6637–6651
Covert SF (1998) Supernumerary chromosomes in filamentous fungi. Curr Genet 33:311–319. https://doi.org/10.1007/s002940050342
Dunham MJ, Badrane H, Ferea T et al (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99:16144–16149. https://doi.org/10.1073/pnas.242624799
Fierro F, Martín JF (1999) Molecular mechanisms of chromosomal rearrangement in fungi. Crit Rev Microbiol 25:1–17. https://doi.org/10.1080/10408419991299185
Forche A, Alby K, Schaefer D et al (2008) The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol 6:e110. https://doi.org/10.1371/journal.pbio.0060110
Fortuna M, Sousa MJ, Côrte-Real M et al (2001) Cell cycle analysis of yeasts. Curr Protoc Cytom 13:11.13.1–11.13.9. https://doi.org/10.1002/0471142956.cy1113s13
Goffeau A, Barrell BG, Bussey H et al (1996) Life with 6000 genes. Science 274:546–567. https://doi.org/10.1126/science.274.5287.546
Gordon JL, Byrne KP, Wolfe KH (2011) Mechanisms of chromosome number evolution in yeast. PLoS Genet 7:e1002190. https://doi.org/10.1371/journal.pgen.1002190
Harari Y, Ram Y, Kupiec M (2018a) Frequent ploidy changes in growing yeast cultures. Curr Genet 64:1001–1004. https://doi.org/10.1007/s00294-018-0823-y
Harari Y, Ram Y, Rappoport N et al (2018b) Spontaneous changes in ploidy are common in yeast. Curr Biol 28:825–835. https://doi.org/10.1016/j.cub.2018.01.062
Hirakawa MP, Chyou DE, Huang D et al (2017) Parasex generates phenotypic diversity de novo and impacts drug resistance and virulence in Candida albicans. Genetics 207:1195–1211. https://doi.org/10.1534/genetics.117.300295
Hull RM, Cruz C, Jack CV, Houseley J (2017) Environmental change drives accelerated adaptation through stimulated copy number variation. PLoS Biol 15:e2001333. https://doi.org/10.1371/journal.pbio.2001333
Iwaguchi S, Homma M, Tanaka K (1992) Clonal variation of chromosome size derived from the rDNA cluster region in Candida albicans. J Gen Microbiol 138:1177–1184. https://doi.org/10.1099/00221287-138-6-1177
Janbon G, Sherman F, Rustchenko E (1998) Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc Natl Acad Sci USA 95:5150–5155. https://doi.org/10.1073/pnas.95.9.5150
Janbon G, Sherman F, Rustchenko E (1999) Appearance and properties of l-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153:653–664
Jeffares DC, Jolly C, Hoti M et al (2017) Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8:14061. https://doi.org/10.1038/ncomms14061
Jeffery-Smith A, Taori SK, Schelenz S et al (2018) Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029. https://doi.org/10.1128/CMR.00029-17
Jones T, Federspiel NA, Chibana H et al (2004) The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA 101:7329–7334. https://doi.org/10.1073/pnas.0401648101
Kabir MA, Ahmad A, Greenberg JR et al (2005) Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc Natl Acad Sci USA 102:12147–12152. https://doi.org/10.1073/pnas.0505625102
Keil RL, Roeder GS (1984) Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39:377–386. https://doi.org/10.1016/0092-8674(84)90016-3
Kobayashi T (2008) A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. Bioessays 30:267–272. https://doi.org/10.1002/bies.20723
Kobayashi T (2014) Ribosomal RNA gene repeats, their stability and cellular senescence. Proc Jpn Acad Ser B 90:119–129. https://doi.org/10.2183/pjab.90.119
Koç A, Wheeler LJ, Mathews CK, Merrill GF (2004) Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279:223–230. https://doi.org/10.1074/jbc.M303952200
Kupiec M (2014) Biology of telomeres: lessons from budding yeast. FEMS Microbiol Rev 38:144–171. https://doi.org/10.1111/1574-6976.12054
Liu J, He MH, Peng J et al (2016) Tethering telomerase to telomeres increases genome instability and promotes chronological aging in yeast. Aging (Albany NY) 8:2827–2847. https://doi.org/10.18632/aging.101095
Lockhart SR, Etienne KA, Vallabhaneni S et al (2017) Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. https://doi.org/10.1093/cid/ciw691
Luo J, Sun X, Cormack BP, Boeke JD (2018) Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 560:392–396. https://doi.org/10.1038/s41586-018-0374-x
Magee BB, Magee PT (1997) WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology 143:289–295. https://doi.org/10.1099/00221287-143-2-289
Maxwell PH (2016) Growth conditions that increase or decrease lifespan in Saccharomyces cerevisiae lead to corresponding decreases or increases in rates of interstitial deletions and non-reciprocal translocations. BMC Genet 17:140. https://doi.org/10.1186/s12863-016-0447-5
Maxwell PH, Burhans WC, Curcio MJ (2011) Retrotransposition is associated with genome instability during chronological aging. Proc Natl Acad Sci USA 108:20376–20381. https://doi.org/10.1073/pnas.1100271108
Measday V, Stirling PC (2016) Navigating yeast genome maintenance with functional genomics. Brief Funct Genom 15:119–129. https://doi.org/10.1093/bfgp/elv033
Mieczkowski PA, Lemoine FJ, Petes TD (2006) Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 5:1010–1020. https://doi.org/10.1016/j.dnarep.2006.05.027
Monerawela C, Bond U (2017) Brewing up a storm: the genomes of lager yeasts and how they evolved. Biotechnol Adv 35:512–519. https://doi.org/10.1016/j.biotechadv.2017.03.003
Muñoz JF, Gade L, Chow NA et al (2018) Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun 9:5346. https://doi.org/10.1038/s41467-018-07779-6
Ni M, Feretzaki M, Li W et al (2013) Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11:e1001653. https://doi.org/10.1371/journal.pbio.1001653
Ormerod KL, Fraser JA (2013) Balancing stability and flexibility within the genome of the pathogen Cryptococcus neoformans. PLoS Pathog 9:e1003764. https://doi.org/10.1371/journal.ppat.1003764
Pal S, Postnikoff SD, Chavez M, Tyler JK (2018) Impaired cohesion and homologous recombination during replicative aging in budding yeast. Sci Adv 4:eaaq0236. https://doi.org/10.1126/sciadv.aaq0236
Pavelka N, Rancati G, Zhu J et al (2010) Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468:321–325. https://doi.org/10.1038/nature09529
Poláková S, Blume C, Zárate JA et al (2009) Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci USA 106:2688–2693. https://doi.org/10.1073/pnas.0809793106
Reedy JL, Floyd AM, Heitman J (2009) Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19:891–899. https://doi.org/10.1016/j.cub.2009.04.058
Rhodes J, Abdolrasouli A, Farrer RA et al (2018) Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 7:43. https://doi.org/10.1038/s41426-018-0045-x
Rustchenko EP, Curran TM, Sherman F (1993) Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol 175:7189–7199. https://doi.org/10.1128/jb.175.22.7189-7199.1993
Rustchenko EP, Howard DH, Sherman F (1994) Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J Bacteriol 176:3231–3241
Rustchenko EP, Howard DH, Sherman F (1997) Variation in assimilating functions occurs in spontaneous Candida albicans mutants having chromosomal alterations. Microbiology 143:1765–1778. https://doi.org/10.1099/00221287-143-5-1765
Sambrook JF, Russell DW (2000) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Satoh K, Makimura K, Hasumi Y et al (2009) Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. https://doi.org/10.1111/j.1348-0421.2008.00083.x
Schelenz S, Hagen F, Rhodes JL et al (2016) First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. https://doi.org/10.1186/s13756-016-0132-5
Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. https://doi.org/10.1126/science.1128242
Selmecki AM, Dulmage K, Cowen LE et al (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5:e1000705. https://doi.org/10.1371/journal.pgen.1000705
Shao Y, Lu N, Wu Z et al (2018) Creating a functional single-chromosome yeast. Nature 560:331–335. https://doi.org/10.1038/s41586-018-0382-x
Sharma C, Kumar N, Meis JF et al (2015) Draft genome sequence of a fluconazole-resistant Candida auris strain from a candidemia patient in India. Genome Announc 3:00722. https://doi.org/10.1128/genomeA.00722-15
Sharma C, Kumar N, Pandey R et al (2016) Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. https://doi.org/10.1016/j.nmni.2016.07.003
Sheltzer JM, Et A (2011) Aneuploidy drives genomic instability in yeast. Science 333:1026–1030. https://doi.org/10.1126/science.1206412
Tang YC, Amon A (2013) Gene copy-number alterations: a cost–benefit analysis. Cell 152:394–405. https://doi.org/10.1016/j.cell.2012.11.043
Todd R, Forche A, Selmecki A (2017) Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution. In: Heitman J, Howlett B, Crous P, Stukenbrock E, James T, Gow N (eds) The fungal kingdom. ASM Press, Washington, DC, pp 599–618. https://doi.org/10.1128/microbiolspec.funk-0051-2016
Torres EM, Sokolsky T, Tucker CM et al (2007) Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916–924. https://doi.org/10.1126/science.1142210
Torres EM, Dephoure N, Panneerselvam A et al (2010) Identification of aneuploidy-tolerating mutations. Cell 143:71–83. https://doi.org/10.1016/j.cell.2010.08.038
Tutaj H, Pogoda E, Tomala K, Korona R (2019) Gene overexpression screen for chromosome instability in yeast primarily identifies cell cycle progression genes. Curr Genet 65:483–492. https://doi.org/10.1007/s00294-018-0885-x
Wendland J, Walther A (2014) Chromosome number reduction in Eremothecium coryli by two telomere-to-telomere fusions. Genome Biol Evol 6:1186–1198. https://doi.org/10.1093/gbe/evu089
Wertheimer NB, Stone N, Berman J (2016) Ploidy dynamics and evolvability in fungi. Philos Trans R Soc B Biol Sci 371:20150461. https://doi.org/10.1098/rstb.2015.0461
Yang F, Kravets A, Bethlendy G et al (2013) Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob Agents Chemother 57:5026–5036. https://doi.org/10.1128/AAC.00516-13
Zhang Y, Ma L-J (2017) Deciphering pathogenicity of Fusarium oxysporum from a phylogenomics perspective. Adv Genet 100:179–209. https://doi.org/10.1016/BS.ADGEN.2017.09.010
Zhang H, Zeidler AFB, Song W et al (2013) Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics 193:785–801. https://doi.org/10.1534/genetics.112.146522
Zolan ME (1995) Chromosome-length polymorphism in fungi. Microbiol Rev 59:686–698
Acknowledgements
We are grateful to Arunaloke Chakrabarti, Anuradha Chowdhary, Elizabeth Johnson (PHE), Takashi Kubota, and Shawn Lockhart (CDC) for providing strains. Flow cytometry was performed at the Iain Fraser Cytometry Centre (IFCC), University of Aberdeen (Raif Yuecel). This work was supported by the Medical Research Council (MRC) Centre for Medical Mycology at the University of Aberdeen [Grant numbers MR/P501955/1, MR/N006364/1]; a Wellcome Trust Institutional Strategic Support Fund grant awarded to the University of Aberdeen [Grant number 204815/Z/16/Z]; a Tenovus Scotland project grant [Grant number G17.02]; a Royal Society Research Grant [Grant number RG140254] awarded to AL; and Wellcome Trust Strategic Award, Senior Investigator and Collaborative Awards [grant numbers 080088, 086827, 075470, 099215, 097377] awarded to NARG.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
No conflict of interest.
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bravo Ruiz, G., Ross, Z.K., Holmes, E. et al. Rapid and extensive karyotype diversification in haploid clinical Candida auris isolates. Curr Genet 65, 1217–1228 (2019). https://doi.org/10.1007/s00294-019-00976-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-019-00976-w