Abstract
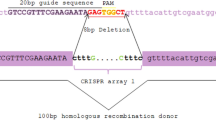
Gene replacement (knock-out) is a major tool for the analysis of gene function. However, the efficiency of correct targeting varies between species, and is dependent on the structure of the DNA construct. We analyzed the targeted insertion mutagenesis method in the budding yeast Saccharomyces castellii, phylogenetically positioned after the whole genome duplication event in the Saccharomyces lineage. We compared the targeting efficiency for target DNA constructs in the respective ends-in and ends-out form. For some of the constructs S. castellii showed a similar high degree of homologous recombination as S. cerevisiae. In agreement with S. cerevisiae, a higher targeting efficiency was seen for the diploid strain than for the haploid. Surprisingly, a higher degree of targeting efficiency was seen for ends-out constructs compared to ends-in constructs. This result may have been influenced by the difference in the length of the homologous target sequences used, although long homology regions of 300 bp–1 kb were used in all constructs. Remarkably, very short regions of cohesive heterologous sequences at the ends of the constructs highly stimulated random illegitimate integration, suggesting that the pathway of non-homologous end joining is highly active in S. castellii.





Similar content being viewed by others
References
Astrom SU, Okamura SM, Rine J (1999) Yeast cell-type regulation of DNA repair. Nature 397:310
Astromskas E, Cohn M (2007) Tools and methods for genetic analysis of Saccharomyces castellii. Yeast 24:499–509
Boulton SJ, Jackson SP (1996) Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J 15:5093–5103
Chan CY, Kiechle M, Manivasakam P, Schiestl RH (2007) Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res 35:5051–5059
Cliften PF, Fulton RS, Wilson RK, Johnston M (2006) After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics 172:863–872
Cohn M (2008) Molecular diversity of telomeric sequences. In: Nosek J, Tomaska L (eds) Origin and evolution of telomeres. Landes Bioscience, Austin, pp 70–82
Cohn M, Blackburn EH (1995) Telomerase in yeast. Science 269:396–400
Cohn M, McEachern MJ, Blackburn EH (1998) Telomeric sequence diversity within the genus Saccharomyces. Curr Genet 33:83–91
Cormack BP, Falkow S (1999) Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151:979–987
Frank-Vaillant M, Marcand S (2002) Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell 10:1189–1199
Hastings PJ, McGill C, Shafer B, Strathern JN (1993) Ends-in vs. ends-out recombination in yeast. Genetics 135:973–980
Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, Friend SH, Marton MJ (2000) Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25:333–337
Kegel A, Sjostrand JO, Astrom SU (2001) Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol 11:1611–1617
Kegel A, Martinez P, Carter SD, Astrom SU (2006) Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res 34:1633–1645
Klinner U, Schafer B (2004) Genetic aspects of targeted insertion mutagenesis in yeasts. FEMS Microbiol Rev 28:201–223
Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38:233–271
Maassen N, Freese S, Schruff B, Passoth V, Klinner U (2008) Nonhomologous end joining and homologous recombination DNA repair pathways in integration mutagenesis in the xylose-fermenting yeast Pichia stipitis. FEMS Yeast Res 8:735–743
Manivasakam P, Weber SC, McElver J, Schiestl RH (1995) Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res 23:2799–2800
Marinoni G, Manuel M, Petersen RF, Hvidtfeldt J, Sulo P, Piskur J (1999) Horizontal transfer of genetic material among Saccharomyces yeasts. J Bacteriol 181:6488–6496
Morrow DM, Connelly C, Hieter P (1997) “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371–382
Naumov GI, Naumova ES, Marinoni G, Piskur J (1998) Genetic analysis of Saccharomyces castellii, S. exiguus and S. martiniae yeasts. Genetika 34:565–568
Orr-Weaver TL, Szostak JW (1983) Multiple, tandem plasmid integration in Saccharomyces cerevisiae. Mol Cell Biol 3:747–749
Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63:349–404
Petersen RF, Langkjaer RB, Hvidtfeldt J, Gartner J, Palmen W, Ussery DW, Piskur J (2002) Inheritance and organisation of the mitochondrial genome differ between two Saccharomyces yeasts. J Mol Biol 318:627–636
Rhodin J, Astromskas E, Cohn M (2006) Characterization of the DNA binding features of Saccharomyces castellii Cdc13p. J Mol Biol 355:335–346
Rothstein RJ (1983) One-step gene disruption in yeast. Methods Enzymol 101:202–211
Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404–409
Svetec IK, Stafa A, Zgaga Z (2007) Genetic side effects accompanying gene targeting in yeast: the influence of short heterologous termini. Yeast 24:637–652
Wach A, Brachat A, Pohlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808
Wahlin J, Cohn M (2002) Analysis of the RAP1 protein binding to homogeneous telomeric repeats in Saccharomyces castellii. Yeast 19:241–256
Wolfe KH (2006) Comparative genomics and genome evolution in yeasts. Philos Trans R Soc Lond B Biol Sci 361:403–412
Acknowledgments
We thank J. Vezilier, H. Mohr, C. Kilgus, W. Su, M. Gsell, I. Baena Ropero, and M. Appelbäck for technical assistance. We are grateful to R.J. Wellinger for sharing the TLC1 sequence data and to U.H. Mortensen for valuable advice. This work was supported by grants from the Carl Trygger Foundation, the Royal Physiographic Society in Lund and the Jörgen Lindström Foundation. E. Astromskas was supported by a scholarship from the Sven and Lilly Lawsky Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Sunnerhagen.
Rights and permissions
About this article
Cite this article
Astromskas, E., Cohn, M. Ends-in vs. ends-out targeted insertion mutagenesis in Saccharomyces castellii . Curr Genet 55, 339–347 (2009). https://doi.org/10.1007/s00294-009-0248-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-009-0248-8