Abstract
A compartment model for an in-host liquid nanoparticle delivered mRNA vaccine is presented. Through non-dimensionalisation, five timescales are identified that dictate the lifetime of the vaccine in-host: decay of interferon gamma, antibody priming, autocatalytic growth, antibody peak and decay, and interleukin cessation. Through asymptotic analysis we are able to obtain semi-analytical solutions in each of the time regimes which allows us to predict maximal concentrations and better understand parameter dependence in the model. We compare our model to 22 data sets for the BNT162b2 and mRNA-1273 mRNA vaccines demonstrating good agreement. Using our analysis, we estimate the values for each of the five timescales in each data set and predict maximal concentrations of plasma B-cells, antibody, and interleukin. Through our comparison, we do not observe any discernible differences between vaccine candidates and sex. However, we do identify an age dependence, specifically that vaccine activation takes longer and that peak antibody occurs sooner in patients aged 55 and greater.
Similar content being viewed by others
1 Introduction
Vaccines are one of the greatest advents of modern society, significantly reducing mortality (cf. Rodrigues and Plotkin 2020) with Ehreth (2003) estimating a prevention of 6 million deaths of vaccine-preventable diseases each year. Vaccines are derived from many precursors including from inactivated virus, viral protein subunits, recombinant human adenovirus, and messenger RNA (mRNA) with the latter being of increasing interest due to their high potency, low manufacturing costs, and the ability to be developed quickly as outlined by Pardi et al. (2018). mRNA vaccines gained prominence during the COVID-19 pandemic with the development and deployment of BNT162b2 (produced by Pfizer-BioNTech) and mRNA-1273 (produced by Moderna) both of which are injected via liquid nanoparticles (LNP). This mechanism was chosen to aid in cell delivery and protect the mRNA from degradation (cf. Ndeupen et al. 2021).
mRNA vaccines have been in development for many years. A review of their usage in infectious diseases has been conducted by Zhang et al. (2019). Prior to COVID-19 it was recognized that mRNA vaccines were outperforming other technologies such as inactivated virus and protein adjuvanted vaccines (cf. Pardi et al. 2018). mRNA vaccines have also demonstrated robust immune responses to other diseases. A study by Bahl et al. (2017) showed vaccines to be effective against severe disease of H7N9 and H10N8 influenza viruses in mice, non-human primates, and ferrets.
mRNA vaccines, as with other vaccine types, demonstrate waning effectiveness. A study by Menni et al. (2022) concluded that antibody protection from COVID-19 mRNA vaccines showed significant waning beginning 5 months after the standard two dose regiment, acknowledging that overall vaccine effectiveness was dependent on age and comorbidity. However, they also saw continued protection from severe disease beyond 6 months. Clinical studies such as these demonstrate the importance of measuring immunity response, development, and decay in mRNA vaccines for COVID-19. However, these trials can be very costly and, without a clear understanding of the immune response, the data collection requirements can be uncertain. Mathematical modelling and analysis provides a cost-effective tool for understanding and predicting the immunity response to vaccines.
Most mathematical modelling of infectious disease occurs at the population scale and throughout the COVID-19 pandemic there have been many such studies (cf. Tang et al. 2020; Moyles et al. 2021; Yuan et al. 2022; Fair et al. 2022; Childs et al. 2022; Vignals et al. 2021; Betti et al. 2021; Dick et al. 2021; Moore et al. 2021; Moss et al. 2020; Smirnova et al. 2021; Wells et al. 2021; Li et al. 2020; Yuan et al. 2022; Hogan et al. 2021). While the impacts of vaccine efficacy and waning are important at these scales, the actual process of immune development occurs within-host. In-host mathematical modelling considers pathogen reproduction and cellular infection within a single individual and has been effectively employed in various diseases. For example, Heffernan and Keeling (2009) modelled vaccination and waning with measles, Herz et al. (1996) modelled the intracellular viral life cycle phase of HIV and hepatitis B, and Perelson (2002) reviewed immune system dynamic modelling for HIV, hepatitis C, and cytomegalovirus (CMV). In-host modelling has been extended to COVID-19 with studies looking at infection (cf. Hernandez-Vargas and Velasco-Hernandez 2020; Perelson and Ke 2021; Kim et al. 2021; Néant et al. 2021; Sadria and Layton 2021; Lin et al. 2022; Ke et al. 2022; Korosec et al. 2023) and vaccination (cf. Farhang-Sardroodi et al. 2021; Korosec et al. 2022; Gholami et al. 2023).
In this paper we explore an in-host mathematical model for an LNP vaccine first considered by Korosec et al. (2022) where computational mixed-effects modelling was used to identify model parameters from a variety of data sets. Their results demonstrated a diverse variability in the parameter estimates and consequently the model comparisons. Furthermore, the nature of the clinical trial data analyzed meant that the time series had coarse resolution spread over a long duration. In this paper, we identify the dominant terms of the model and the timescales over which they are relevant with the goal of elucidating the antibody process and providing direction for improved data resolution. However, we also provide mechanistic insight into the immune response which is important because the precise timescales of activating the immune response in humans is relatively unknown. Luceripherase mouse studies have been conducted, for example, by Pardi et al. (2015) and Lutz et al. (2017) showing that the injection site remains active for approximately 7–10 days and that luceriphase was also present in the draining lymph nodes nearest the injection site a few days after injection. Similar luceriphase studies cannot be done on humans and instead highly resolved blood draws are needed to monitor the adaptive and innate immune response. Our timescale analysis provides a framework to understand these physiological processes sequentially.
Understanding a sequential ordering of processes that drive vaccine dynamics is very beneficial. It helps understand and separate the innate immune responses from the adaptive ones generated by the vaccine. Separating these timescales helps identify balance between the immune responses which is important for limiting the body’s ability to attack vaccine species at dosage. A thorough model analysis also improves mechanistic understanding. Even though mRNA vaccines have demonstrated strong immune responses, the immunological activity leading to these responses is less clear (cf. Lindgren et al. 2017). Our analysis generalizes beyond mRNA vaccines as well. The model we present is itself adapted from an adenovirus-based vaccine. Therefore, our work can be used to broadly decompose vaccine-mediated immunity for a variety of candidates and diseases.
A common approach in immunological modelling studies is to focus solely on simulations. Our approach exploits asymptotic methods to isolate model structure and function as it evolves with time identifying important parameter relationships. While this methodology is mathematically richer than solely a simulation based study, it can also provide practical and biological insight. For example, parameter identifiability is an important consideration in modelling studies. Analytic expressions determine explicit relationships with parameters which can elucidate both model identifiability, i.e., which parameters form scaling groups, as well as practical identifiability, i.e., what time points are needed in data collection to identify certain parameters. Formal asymptotic methods can justify the inclusions or removal of certain terms in a model adding rigor and confidence to biological intuition. Analytic methods can enrich biological understanding of a problem by linking processes with explicit parameter relationships. For example, understanding a peak immunological response explicitly in terms of model parameters can provide insight into the processes that drive this response while also allowing researchers to exploit new vaccines and therapies to improve the response.
The paper is organized as follows. We introduce and summarize the model of Korosec et al. (2022) in Sect. 2. We nondimensionalize the model in Sect. 2.1 where, through some assumption of scales, we are able to reduce the model to a simpler form. We identify a series of timescales in Sect. 3 that account for antibody development, growth, maximum, and ultimately decay. In each timescale regime we are able to determine asymptotic analytic solutions that explain the immunity response. We compare these analytic results to a full numerical simulation of the model in Sect. 4 where maxima of key immunity factors are approximated and we compare the model to data sets from a series of clinical trials. We discuss results and conclude the paper in Sect. 5.
2 Model summary and reduction
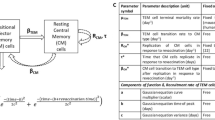
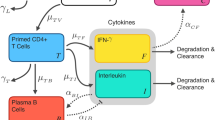
We consider a compartmental ordinary differential equation model first published by Farhang-Sardroodi et al. (2021) for adenovirus-based vaccines and later adapted by Korosec et al. (2022) for LNP mRNA vaccines. The process being modelled begins with the injection of a concentration of LNP (L) that diffuses through inactive target cells of which we assume there is an infinite reservoir. Details of these target cells are unclear. When injected intramuscularly, such as with humans, Lutz et al. (2017) found in a study of mice that mRNA vaccines activated the innate immune system near the site of injection as well as in draining lymph nodes. These target cells activate to become vaccinated cells (V) that promote the production of CD4\(^+\) (T) and cytotoxic CD8\(^+\) (C) T-cells. These CD4\(^+\) T-cells further promote the production of plasma B-cells (B), interferon-\(\gamma \) (IFN-\(\gamma \), F), and interleukin (I). Finally, the plasma B-cells stimulate production of immunoglobulin G (IgG) antibody (A). The equations of the model studied by Korosec et al. (2022) are given by
The parameters \(\mu _{ij}\) are priming rates (\(\hbox {d}^{-1}\)) that activate component j from component i. \(\beta _{ij}\) are immune response autocatalytic rates (\(\hbox {d}^{-1}\)) arising from non-linear activation and inhibition of species j on species i, and \(\gamma _i\) are the natural decay rates (\(\hbox {d}^{-1}\)). The parameters \(s_\textrm{I}\) and \(s_\textrm{F}\) are concentrations of interleukin and IFN-\(\gamma \), respectively, that produce half-maximal autocatalytic rates, \(\beta _{ij}\). Time, t, is measured in days and concentrations of each component are in arbitrary volumetric units. We note that equations (1d) and (1f) contain saturation dynamics through the terms \(s_F\) and \(s_I\). We include these to be consistent with the full model of Korosec et al. (2022), but biologically it will be important to limit activating kinetics in a general model of vaccination which could include multiple doses.
The only feasible steady state to (1) is every concentration being zero and thus, in the absence of some initial vaccine, it is reasonable within the context of the model to measure no immune response. However, it is possible through previous infection or other biological mechanisms not accounted for in the model, that some basal concentrations may exist for quantities not primed through vaccine. As such, we consider the generalized initial conditions to (1) to be
where we have assumed that vaccine is injected at time \(t=0\) with concentration \(L_\textrm{i}\).
2.1 Non-dimensionalization and model reduction
To understand the effects of each term in the model we non-dimensionalize (1). Firstly, the concentration of LNP will be determined by its initial value as it only decays and thus we scale \(L\sim L_\textrm{i}\). Secondly, as the dynamics are driven by vaccination, the natural time scale is the activation of vaccinated cells by LNP and thus we scale \(t\sim \mu _\textrm{LV}^{-1}\). We do not scale other variables by their initial condition because we expect significant variability in these levels between individuals. Furthermore, since the model is derived based on vaccine response, we anticipate that it should be driven by the dynamics of vaccination rather than any latent initial concentrations present. Instead, we look to (1) and choose scales by balancing source and sink terms in the model. For example in (1b), (1c), (1d), (1f), and (1h) vaccine priming drives production and so it is sensible to choose scales for V, T, B, C, and I driven by the source terms. For IgG in (1e) it is sensible to balance the source of plasma cells with decay of antibody, A. For F in (1g), we scale the concentration of IFN-\(\gamma \) assuming its natural decay is the dominant sink term. This is consistent with fast decay rates of IFN-\(\gamma \) of between 3 and 40 min in mice and humans reported by Foon et al. (1985); Gonias et al. (1988); Psimadas et al. (2012). We note that this, and all decay terms, strictly refers to the removal from the vaccine induced antibody response being modelled here. It does not necessarily mean that the components are degrading, but could instead by absorbed or consumed in other biological functions not related to immune response of the vaccine. Overall, we are led to the following scales for each of the concentrations
which leads to the non-dimensional model
where dot indicates differentiation with respect to non-dimensional time. The parameters in (4) are defined as follows:
Many of these parameters have natural interpretations. For example \(\epsilon \) is the timescale ratio of vaccine absorption to decay of IgG, the terminal antibody; the parameter \(\alpha _{i}\) is the timescale ratio of the natural decay of concentration i relative to the decay of IgG; and the parameter \(\delta \) is the timescale ratio of release of vaccine to decay of IFN-\(\gamma \). The parameters \(\kappa _\textrm{I}\) and \(\kappa _\textrm{F}\) are effective saturation constants which limit the production of plasma B cells and cytotoxic T-cells while the parameters \(\lambda _{i}\) are the strengths of the autocatalytic production terms compared to the vaccine priming sources for the B, C, F, and I compartments. Each of \(\mathcal {A}\), \(\mathcal {F}\), and \(\mathcal {I}\) are non-dimensional initial concentrations of IgG, IFN-\(\gamma \), and interleukin respectively.
We anticipate that an effective immune response necessitates that the decay of antibodies is much slower than the absorption of vaccine in the LNP and thus that \(\epsilon \ll 1\). This is supported experimentally where Lutz et al. (2017), for example, observed that mice began to produce antigen at the site of injection on the order of hours after the needle, while antibody decay halflife was on the order of many days. Conversely, because of the fast decay rates on the order of minutes of IFN-\(\gamma \) observed by Foon et al. (1985) and others compared to the LNP absorption timescale of hours observed by Lutz et al. (2017), we will assume that \(\delta \ll 1\) as well. We anticipate that the ratio of the remaining decay rates to \(\gamma _A\) are comparable and thus take \(\alpha _i\sim \mathcal {O}(1)\). Without any presupposition about the magnitude of production multipliers, \(\lambda _{i}\), we simply assume formally that they are \(\mathcal {O}(1)\). So long as terms are not an order of magnitude larger than we have assumed, the model will be consistently scaled and its structure and analysis will be valid. The actual size of terms is dependent on specific case studies, a detail we resolve in Sect. 4.2.
Assuming the saturation parameters \(\kappa _i\) are small is generally associated with limited saturation effect. While this is likely true in (4g) where IFN-\(\gamma \) decays quickly, it is less clear in (4d) where autocatalytic production of interleukin may significantly contribute to antibody production. However, if saturation effects become important then this is likely a consequence of an overactive immune system and we assume that is generally not the case.
Based on the assumptions, we propose a reduced model to (4) where \(\kappa _\textrm{I}=\kappa _\textrm{F}=0\). Furthermore, since both \(\epsilon \ll 1\) and \(\delta \ll 1\) then the terms associated to \(\lambda _\textrm{C}\) in (4f) and \(\lambda _\textrm{F}\) in (4g) are likely non-identifiable and therefore, we will assume that \(\lambda _\textrm{F}=\lambda _\textrm{C}=0\) without loss of generality. Finally in (4a) since we have two competing sink terms we assume the second, being the smaller of the two, is non-identifiable. Thus we will assume that \(\alpha _\textrm{L}=0\). Overall, we are led to the following reduced model:
subject to the same initial conditions as (4). It may seem unsatisfactory that we have neglected some small terms in the reduced model (6) while leaving others. This is justified from (6e) where if we take \(\epsilon \equiv 0\) there is no mechanism for antibody response. Therefore, necessarily, there must be some long timescale antibody production driven by \(\epsilon \). Thus, we leave the small terms associated to this parameter. We note that this is also consistent with not scaling terms by latent initial conditions. We demonstrate agreement between the full model (4) and the reduced model (6) in Sect. 4.
3 Timescale decomposition
We will now analyze the reduced model (6) to understand the impact of different parameters on the long-term antibody response. We begin by noting that L, V, T, and C in (6) can be solved analytically as these are a cascade of linear equations that decouple from one another. The solutions, subject to the appropriate initial conditions \(L(0)=1\) and \(V(0)=T(0)=C(0)\), are
We note in model (7) we have made the explicit assumption that \(\alpha _\textrm{T}\ne \alpha _\textrm{V}\) and \(\alpha _\textrm{C}\ne \alpha _\textrm{V}\) for generality. We will use the assumption that all \(\alpha _\textrm{V}\), \(\alpha _\textrm{T}\), and \(\alpha _\textrm{B}\) are unique throughout the remainder of the manuscript and discuss the specific case where the parameters are equal in Appendix B.
The anti-body concentration, (6e) depends on the plasma B-cell concentration, B, and thus can not yet be solved in closed form. However, we can write its solution in terms of the plasma B-cell concentration,
We note that we can also solve (6g) which has the general solution,
with T given by (7c). However, aside from a fast timescale of \(\mathcal {O}(\delta )\) as described in Sect. 3.1, this equation is mostly in quasi-steady state with
which is more insightful than the full solution (9).
The solutions (7), (8), and (9) already significantly decouple the eight equation model (6) to a two equation model for B, and I which we now systematically explore through a series of chronological time scales. The full description of these timescales is presented and derived in Sects. 3.1 to 3.5, but are also summarized in Table 1 for brevity.
3.1 IFN-\(\gamma \) clearance
The first time scale emerges in (6g) when \(t\sim \mathcal {O}(\delta )\), recalling that \(\delta \) is the ratio of vaccine absorption to IFN-\(\gamma \) decay. As such we let \(t=\delta t_1\) resulting in every equation of (6) to effectively be in equilibrium except for (6g) which becomes
The CD4\(^+\) T-cells at this timescale are given by (7c) after substituting \(t=\delta t_1\) and expanding for \(\delta \ll 1\). This results in \(T\approx 0\) and thus that
Therefore, at this timescale we have that any initial concentration of IFN-\(\gamma \) quickly clears the body. Conversely, if there is no initial concentration of IFN-\(\gamma \) then this timescale can be ignored as none of the other components have yet activated, since B and I are given by their initial conditions to leaing order when \(t\sim \mathcal {O}(\delta )\).
3.2 Vaccine priming
The next time scale occurs when \(t\sim \mathcal {O}(1)\), i.e. the selected timescale from the non-dimensionalisation. Having already removed terms via (7) and (8), and recalling that \(F=T\) from the quasi-steady limit (10), the reduced model at this scale is
These equations are non-linear and do not have direct analytic solutions. Exploiting the smallness of the parameter \(\epsilon \) we can expand (7c) when \(t\sim \mathcal {O}(1)\) and \(\epsilon \ll 1\) to get,
for the CD4\(^+\) T-cell population at this timescale. The formulation of this asymptotic series suggests we pose expansions \(B=B_0+\epsilon B_1\) and \(I=I_0+\epsilon I_1\) and substitute into (12). The leading order problem becomes
subject to \(B_0(0)=0\) and \(I_0(0)=\mathcal {I}\) which has solution
Expanding (12) to \(\mathcal {O}(\epsilon )\) we get
which has solution
where we define
Having determined B, the anti-body concentration is given by expanding (8) for \(t\sim \mathcal {O}(1)\) and \(\epsilon \ll 1\) which yields,
The leading order anti-body response is given by its initial condition and thus there is very little production at the \(t\sim \mathcal {O}(1)\) timescale. This result supports the physiological intuition that, at the \(\mathcal {O}(1)\) timescale, the plasma B-cells and interleukin have yet to develop. Thus, antibodies are close to their background levels during this timescale.
The need for a longer time scale beyond \(t\sim \mathcal {O}(1)\) is immediately clear as each of the concentrations T, B, A, and I grow without bound. From (13), T loses asymptotic consistency when \(t\sim \mathcal {O}(\epsilon ^{-1})\) because the correction term grows quicker than the leading order term. This represents a time where the production of CD4\(^+\) T cells from the vaccine has stopped and only decay of remaining cells is taking place. However, from (17a) the plasma B-cell concentration loses asymptotic consistency when \(t\sim \mathcal {O}(\epsilon ^{-1/3})\), an earlier time than \(t\sim \mathcal {O}(\epsilon ^{-1})\). This breakdown is also evident in (17b) for interleukin and (18) for IgG antibody.
The early breakdown of the solution to the plasma B-cell concentration is due to the rapid autocatalytic production of plasma B-cells as interleukin concentration increases. Therefore, \(t\sim \mathcal {O}(\epsilon ^{-1/3})\) represents a switchover from the vaccine production of plasma B-cells being the dominant contribution to the self-stimulating autocatalytic production becoming dominant. We find that this breakdown happens beyond the leading order dynamics necessitating the two-term asymptotic expansion posed at this timescale.
3.3 Antibody autocatalytic growth
We introduce the new timescale \(t=\epsilon ^{-1/3}{t_{{3}}}\), substitute into (7c) and take \(\epsilon \ll 1\) to get that the T-cell concentration at this scale is,
where we have scaled \(T=\epsilon ^{-1/3}{T_{{3}}}\). Unsurprisingly, we see that \({T_{{3}}}\) loses asymptotic consistency when \({t_{{3}}}\sim \mathcal {O}(\epsilon ^{-2/3})\) equivalent to the asymptotic breakdown when \(t\sim \mathcal {O}(\epsilon ^{-1})\) as discussed in Sect. 3.2. Substituting the \(t_3\) timescale into (15b) and (15a) shows that it is sensible to scale \(I=\epsilon ^{-2/3}{I_{{3}}}\) and \(B=\epsilon ^{-2/3}{B_{{3}}}\). This leads to the reduced model (6) at this scale
Unlike the \(\mathcal {O}(1)\) time in Sect. 3.2, the leading order solution will be sufficient for capturing the dynamics at this timescale (see Appendix A) and this leading order is given by
We obtain the leading order antibody concentration by substituting \(t=\epsilon ^{-1/3}{t_{{3}}}\) into (8) and expanding for \(\epsilon \ll 1\) yielding,
Multiplying (21a) by \(\lambda _\textrm{I}\) and (21b) by \(\lambda _\textrm{B}\) and adding yields
and thus
where we have chosen the integration constant so that both \({B_{{3}}}\) and \({I_{{3}}}\) tend to \({t_{{3}}}^2/2\) as required for matching to (15a) and (15b) when \({t_{{3}}}\ll 1\). Substituting (24) into (21a) yields
This non-linear equation can be solved using Kummer functions (see Appendix A) leading to the solution
with \(\text {M}(x,y,z)\) and \(\text {U}(x,y,z)\) Kummer functions of the first and second kind respectively and \(\Gamma (z)\) the usual Gamma function (cf. Abramowitz and Stegun 1983; Polyanin and Zaitsev 2017). The leading order solution (26) satisfies the correct matching condition that \(B_{3_0}\sim {t_{{3}}}^2/2\) for \({t_{{3}}}\ll 1\) see (A9) in Appendix A for details coming from the far-field behaviour of the solution (15a) in the \(t\sim \mathcal {O}(1)\) timescale. Having determined \(B_{3_0}\) then \(I_{3_0}\) is determined from (24).
We show in Sect. 1 of Appendix A that the solutions at this timescale do not lose asymptotic consistency prior to \(t\sim \mathcal {O}(\epsilon ^{-1})\) when the CD4\(^+\) T-cell concentration given by (19) loses asymptotic consistency and therefore there are no additional intermediate timescales for antibody growth to consider. This resolves the loss of asymptotic consistency for B, I, and A that was discovered at the \(t\sim \mathcal {O}(1)\) timescale in Sect. 3.2. However, the loss of asymptotic consistency in the CD4\(^+\) T-cell concentration is still unresolved as that occurred when \(t\sim \mathcal {O}(\epsilon )\) due to the decay terms not being included.
3.4 Peak and decay of antibody
Introducing the timescale \(t=\epsilon ^{-1}t_4\), we expand (7c) for T yielding,
where we have scaled \(T=\epsilon ^{-1}T_4\). In Appendix A we show the behaviour of B, A, and I when \({t_{{3}}}\gg 1\) given by (A20), (A17) and (A21) respectively. This provides the matching scaling at the \(t_4\) timescale and therefore suggests we scale \(B=\epsilon ^{-2}B_4\) and \(I=I_4\) transforming the reduced model (6) at this scale to
From (28b) we have that \(I_4\) is in a quasi-steady state up to \(\mathcal {O}(\epsilon ^2)\) and therefore,
to leading order. As with the \(t_3\) timescale in Sect. 3.3, the leading order problem will be sufficient to capture the dynamics in this \(t_4\) timescale. Substituting (29) into (28) yields
which has solution
We note that for \(t_4\ll 1\) that
which matches \({B_{{3}}}\) for \({t_{{3}}}\gg 1\) as required (see (A16) in Appendix A). Following (8), after taking \(t=\epsilon ^{-1}t_4\) and scaling \(A=\epsilon ^{-2}A_4\), then the antibody concentration satisfies,
to leading order.
Since we have assumed that \(\alpha _\textrm{V}\), \(\alpha _\textrm{T}\) and \(\alpha _\textrm{B}\) have unique values, for \(t_4\gg 1\),
where \(\alpha _i=\text {min}(\alpha _\textrm{V},\alpha _\textrm{T})\) and \(\alpha _j=\text {max}(\alpha _\textrm{V},\alpha _\textrm{T})\). Similarly for plasma B-cells,
where \(\hat{\alpha }_i=\text {min}(\alpha _\textrm{V},\alpha _\textrm{T},\alpha _\textrm{B})\), \(\hat{\alpha }_k=\text {max}(\alpha _\textrm{V},\alpha _\textrm{T},\alpha _\textrm{B})\), and \(\hat{\alpha }_i<\hat{\alpha }_j<\hat{\alpha }_k\). If \(\hat{\alpha }_i=\alpha _\textrm{B}\) then from (29),
If instead \(\alpha _\textrm{B}>\hat{\alpha }_i\) then \(\hat{\alpha }_i=\alpha _i\) and
which is a constant. However, if we look at the full equation (28b) then the steady state interleukin concentration, \(I_4^\infty \), is
because \(T_4^\infty =B_4^\infty =0\). Therefore if T and B decay at the same rate then a final timescale emerges to resolve the decay of interleukin.
3.5 Interleukin cessation
We assume for this Section that \(\alpha _\textrm{B}>\hat{\alpha }_i\) so that (37) is in disagreement with the steady state interleukin (38) when \(t_4\gg 1\) necessitating the new timescale. Physiologically, this will occur when plasma B-cells do not decay slower than CD4\(^+\) T-cells. The failure of interleukin decay stems from neglecting the \(\alpha _\textrm{I}\) term in the denominator of (38) which cannot be done if \(B_4\sim \mathcal {O}(\epsilon ^2)\). From (35) this occurs approximately when \(\text {e}^{-\alpha _it_4^*}=\epsilon ^2\) or \(t_4^*=-\tfrac{2}{\alpha _i}\log \epsilon \) and so we introduce a fifth and final timescale, \(t=\epsilon ^{-1}(t_4^*+t_5)=\epsilon ^{-1}(-\tfrac{2}{\alpha _i}\log \epsilon +t_5)\) with the additive form of \(t_5\) originating from the exponential decay behaviour in B. We substitute this timescale into (7c) for T to yield,
to leading order. The reduced model (6) at this order becomes a fully non-linear coupled model which cannot be solved analytically. However, from (35) we know that \(B_4\) is decaying exponentially for \(t_4\gg 1\). The introduction of the \(t_5\) timescale was to include the natural decay of interleukin and as such we will approximate the plasma B-cell concentration with (35) substituting in the \(t_5\) timescale,
With this approximation, the antibody response can also be continued from (33).
Using (39) and (40) allows simplification of the reduced model (6) to a linear equation for interleukin,
with solution,
We note that in (42) we have used \(I_{5_0}=(\alpha _\textrm{B}-\alpha _i)(\lambda _\textrm{B}+\lambda _\textrm{I})^{-1}\) when \(t_5=0\) following (37).
The \(t_5\) timescale captures the final decay of interleukin and thus completes the dynamics of the model.
4 Results
We begin by demonstrating that the five timescales discussed in Sect. 3 reasonably capture the model dynamics. We plot L, V, T, C, and F, which can be determined analytically for all time, in Fig. 1. We plot B, A, and I, which required asymptotic decomposition, in Fig. 2. We simulate both the full model (1) and the reduced model (6) to demonstrate the negligible effect of ignoring the parameters \(\lambda _\textrm{F}\), \(\lambda _\textrm{C}\), \(\alpha _\textrm{L}\), and \(\kappa _i\). We have chosen \(\alpha _\textrm{B}>\alpha _\textrm{V}>\alpha _\textrm{T}\) so that interleukin cessation dynamics can be observed. To indicate the different timescales described in Sect. 3 we alternate a gray-white background with the first gray background being the \(t_1\) range, the first white background being the \(t\sim \mathcal {O}(1)\) range, the next gray background being the \(t_3\) range etc. We plot the time on a logarithmic axis to capture the full range of dynamics from \(\mathcal {O}(\delta )\) to \(\mathcal {O}(\epsilon ^{-1})\). In Fig. 2, for the asymptotic expansion comparison, we use only the leading order terms that were derived in Sect. 3. The one exception to this is the \(t_2=t\sim \mathcal {O}(1)\) timescale in Sect. 3.2 where a two-term expansion was computed to show the loss of asymptotic consistency. Therefore, we use the full two-term expansion for comparison.
Comparison of the full model (4), the reduced model (6), and the analytic approximations to L, V, T, C, and F given by (7) and (10) respectively with parameters \(\epsilon =\delta =\kappa _\textrm{F}=\kappa _\textrm{I}=0.01\), \(\alpha _\textrm{L}=\alpha _\textrm{T}=\alpha _\textrm{C}=\alpha _\textrm{I}=\lambda _i=1\), \(\alpha _\textrm{V}=2\), and \(\alpha _\textrm{B}=3\) on a logarithmic non-dimensional time series. For all simulations the initial conditions are \(L(0)=A(0)=F(0)=I(0)=1\) and \(V(0)=T(0)=B(0)=C(0)=0\). We alternate the plot with gray and white patches to indicate the regions where each timescale is valid. Since we have taken \(\epsilon =0.01\) then the start of each time interval is \(t_1=0.01\), \(t_2=t=1\), \(t_3=4.64\), \(t_4=100\), and \(t_5=921.03\) which are non-dimensional times. For F we note the red-dashed curve represents that quasi-steady limit \(F=T\) derived for all timescales beyond \(t_1\) as detailed in Sect. 3
Comparison of the full model (4), the reduced model (6), and the asymptotic approximations to B, A, and I described in Sect. 3 with parameters \(\epsilon =\delta =\kappa _\textrm{F}=\kappa _\textrm{I}=0.01\), \(\alpha _\textrm{L}=\alpha _\textrm{T}=\alpha _\textrm{C}=\alpha _\textrm{I}=\lambda _i=1\), \(\alpha _\textrm{V}=2\), and \(\alpha _\textrm{B}=3\) on a logarithmic non-dimensional time series. For all simulations the initial conditions are \(L(0)=A(0)=F(0)=I(0)=1\) and \(V(0)=T(0)=B(0)=C(0)=0\). We alternate the plot with gray and white patches to indicate the regions where each timescale is valid. Since we have taken \(\epsilon =0.01\) then the start of each time interval is \(t_1=0.01\), \(t_2=1\), \(t_3=4.64\), \(t_4=100\), and \(t_5=921.03\)
We observe excellent agreement between simulation and analytic results in Fig. 1. Since we have chosen a non-zero initial condition for IFN-\(\gamma \), in Fig. 2 the initial condition decays to zero in the \(t_1\) timescale before reaching the \(F=T\) quasi-steady value as discussed in Sect. 3.1. Figure 2 showcases the role of the autocatalytic timescale \({t_{{3}}}\) discussed in Sect. 3.3. The plasma B-cell and antibody response in Fig. 2a, b, respectively, do not have a substantial region in the \({t_{{3}}}\) timescale where the asymptotic solutions compare favourably with the numerical ones. Instead, the solution transfers smoothly from the \(t\sim \mathcal {O}(1)\) timescale (discussed in Sect. 3.2) to the \(t_4\) timescale (discussed in Sect. 3.4). However, as seen in Fig. 2c, the interleukin dynamics are highly captured by the asymptotic solution in the \(t_3\) timescale and its maximal concentration is reached within the \(t_3\) region. Interleukin plays an activator-inhibitor role in the system. It activates the plasma-B cells to initiate the autocatalytic production but inhibits itself which ultimately terminates the autocatalytic production. The maximal value of interleukin represents a shift from the activator to inhibitor role. As observed in Fig. 2c the maximum occurs quite early in the \(t_3\) timescale explaining why B and A quickly follow the dynamics at the \(t_4\) timescale described in Sect. 3.4. We emphasize that the solutions in each timescale are formally valid for an arbitrary small parameter, \(\epsilon \), for the regions they are defined. When choosing an actual value for \(\epsilon \), such as \(\epsilon =0.01\) in Fig. 2 then the asymptotic structure can breakdown numerically. This is particularly important for the autocatalytic region described in Sect. 3.3 which occurs at an order \(\epsilon ^{-1/3}\). Formally, \(\epsilon ^{-1/3}\gg 1\), however, if \(\epsilon =0.01\), for example then \(\epsilon ^{-1/3}=4.6\) which is not particularly large. In this timescale we have \(I\sim \mathcal {O}(\epsilon ^{-2/3})\approx 21\) when \(\epsilon =0.01\). As can be seen in Fig. 2c and is detailed in Sect. 4.1, interleukin transitions through its maximum throughout the \(t_3\) timescale and so very quickly I returns to \(\mathcal {O}(1)\) practically entering the \(t_4\) regime. Thus, numerical realizations of \(\epsilon \) may cause the solutions in timescales \(t_2\), \(t_3\), and \(t_4\) to be appear to be valid longer (or shorter) than theoretically predicted in the analysis. We could improve the asymptotic agreement by considering the composite solution which is the sum of the different asymptotic solutions with their overlapping contributions removed, but omit the details here.
4.1 Maximal concentrations
The asymptotic solution for interleukin and plasma B-cells at the \(t_3\) timescale provides us a mechanism to determine the maximal interleukin concentration. At the maximal concentration, \({I_{{3}}}_{{t_{{3}}}}=0\), and thus from (23) that \({B_{{3}}}_{{t_{{3}}}}=\lambda _\textrm{I}^{-1}(\lambda _\textrm{I}+\lambda _\textrm{B}){t_{{3}}}\) to leading order. Substituting this into the differential equation for \({B_{{3}}}_0\) (25) yields,
where \(\hat{{t_{{3}}}}\) is the time at which the maximal interleukin concentration occurs and we define \(\hat{{B_{{3}}}}={B_{{3}}}(\hat{{t_{{3}}}})\). Solving (43) for the plasma B-cell concentration yields,
This expression only has a solution so long as \(\hat{{t_{{3}}}}\) is large enough, namely
Prior to this time, \({B_{{3}}}\) cannot grow fast enough to generate an optimal value for the interleukin concentration. Therefore, while (45) does not provide any insight into the value for \(\hat{{t_{{3}}}}\), it does provide an estimate for the minimum time before interleukin begins inhibiting antibody production which is useful information for understanding the dynamics of the immune response. To actually estimate the value for \(\hat{{t_{{3}}}}\), it is more useful to solve (43) for \(\hat{{t_{{3}}}}\). This yields,
Using (45) as an initial guess, we can compute \(\hat{{B_{{3}}}}\) from (26). Substituting this value into (46), we update \(\hat{{t_{{3}}}}\). We then iterate until we converge to a desired tolerance. For the parameters in Fig. 2c, we have that \(\hat{{t_{{3}}}}=1.714\) and \({I_{{3}}}(\hat{{t_{{3}}}})=0.8035\). Taking scales into account, these correspond to non-dimensional time \(t_{I_\textrm{max}}=7.953\) and concentration \(I_{I_\textrm{max}}=17.31\) and are comparable to the maximal values numerically obtained from simulating the full model (4), \(t_{I_\textrm{max}}^\textrm{sim}=9.227\) and \(I_{I_\textrm{max}}^\textrm{sim}=17.25\) respectively.
The maximum for plasma B-cells occurs in the \(t_4\) timescale. From (30), the maximum B concentration to leading order occurs when
From (27) and (31) this happens when \(t_4=\hat{t}_{4B}\), occurring when
Recall that \(\alpha _\textrm{V}\), \(\alpha _\textrm{T}\), and \(\alpha _\textrm{B}\) are ordered such that \(\hat{\alpha }_i<\hat{\alpha }_j<\hat{\alpha }_k\). The expression (48) is a polynomial equation in \(\text {e}^{-\hat{\alpha }_i\hat{t}_{4B}}\) and thus can be solved in a straight forward way. From the parameters in Fig. 2a we have that (48) gives \(\hat{t}_{4B}=\log (3)\) and \(B_4(\hat{t}_{4B})=\tfrac{4}{27}\). Accounting for scaling, these correspond to non-dimensional time \(t_{B_\textrm{max}}=\log (3)\epsilon ^{-1}=109.9\) and concentration \(B_{B_\textrm{max}}=\tfrac{4}{27\epsilon ^2}=1.483\times 10^{3}\) and compare favourably to the numerical values from the simulation of the full model (4) given by \(t_{B_\textrm{max}}^\textrm{sim}=111.3\) and \(B_{B_\textrm{max}}^\textrm{sim}=1.467\times 10^{3}\). Interestingly, we find that the leading order maximal time given by (48) is independent of the parameters \(\lambda _\textrm{B}\) and \(\lambda _\textrm{I}\), depending only on the decay parameters.
The maximum for IgG antibody also occurs in the \(t_4\) timescale and from (6e) occurs at a time \(\hat{t}_{4A}\) when \(A_4=B_4\) (with \(A_4\) given by (33) and \(B_4\) given by (31)). Due to the integration in the solution for \(A_4\), this may not be an exponential polynomial (such as is the case with the parameters in Fig. 2). However, it can still be solved with a straightforward root-finding process. This yields the time of the maximum antibody concentration for the parameters in Fig. 2b as \(\hat{t}_{4A}=1.973\) with concentration \(A_4(\hat{t}_{4A})=0.1031\). Accounting for scales, these values are \(t_{A_\textrm{max}}=197.3\) and \(A_{A_\textrm{max}}=1.031\times 10^{3}\) for non-dimensional time and concentration, which compare well to the numerical maximal values from simulating the full model (4), \(t_{A_\textrm{max}}^\textrm{sim}=198.6\) and \(A_{A_\textrm{max}}^\textrm{sim}=1.021\times 10^{3}\). Since the antibody is determined as an integral of the plasma B-cells, its time of maximum concentration also does not depend on the growth parameters \(\lambda _\textrm{B}\) and \(\lambda _\textrm{I}\).
4.2 Comparison to data
Parameter fits to the full model (1) for 22 different data sets were performed by Korosec et al. (2022) using mixed-effects modelling software Monolix. The data available was for IgG antibodies, interleukin, and IFN-\(\gamma \) from patients receiving two doses of SARS-CoV-2 vaccine produced by Pfizer-BioNTech (BNT162b2) or Moderna (mRNA-1273). Each data point refers to a time point measurement of either antibody, IFN-\(\gamma \), or interleukin for a given patient. Since each study has multiple patients, some studies have multiple measurements at a given time. Patients are distinct between studies, but within each study,patients are followed chronologically through time taking additional measurements at different time points. However, these individual patient trajectories cannot be identified from the aggregate data. Therefore, for the purpose of parameter fitting in Korosec et al. (2022), “individual” refers to all of the data in a single data set and “population” refers to the collection of 22 data sets. For all details about parameter estimates including inferred parameter distributions see Korosec et al. (2022) and the supplementary material therein.
Using the fit dimensional parameters from Korosec et al. (2022), we compute the equivalent non-dimensional parameters in Table 2 where we also include the average values for each of the two vaccines considered. We bold values in Table 2 that violate the model assumption of parameters being \(\mathcal {O}(1)\) or smaller. The scales for each of the vaccine model compartments given by (3) as well as the non-dimensional non-zero initial values \(\mathcal {A}_0\) and \(\mathcal {I}_0\) are computed in Table 3 for each of the parameter sets (an initial condition \(\mathcal {F}=0\) was taken for all data). We compute the timescales discussed in Sect. 3 for each of the data sets in Table 4 along with the number of data points that occur within each timescale. The number of data points is important because it provides a measure of data resolution and the ability to capture all the dynamics of the immune response.
The number of data points in Table 4 is slightly misleading. Many data sets have multiple patients and therefore much of the data is taken at a single time point for multiple people rather than as a time series for a single person. Secondly, the number of data points in \(0\le t<t_1\) (first column of bold values) are all initial values taken just after vaccination (day 0) and do not actually capture any of the fast IFN-\(\gamma \) dynamics discussed in Sect. 3.2. The model analysis predicts that the majority of antibody activity occurs within the \(t_3\) and \(t_4\) timescales and this is precisely where the majority of data points are collected. Furthermore, we remark the onset of the \(t_4\) timescale which is the timescale over which antibody concentrations peak and decay aligns remarkably well with the timing of the second dosage given as the last column of Table 4.
Table 5 calculates an estimation of the maximal concentration and times they occur for interleukin, plasma B-cells, and IgG antibody following the discussion in Sect. 4.1. The sequence of timescales discussed in Sect. 3 indicates that interleukin reaches maximal value first (in the \(t_3\) timescale) which triggers the inhibition process of plasma B-cells leading to its maximal value (in the \(t_4\) timescale) and then the maximal value of antibody followed shortly. Thus, generally we suspect an ordering of \(t_{I_{\max }}<t_{B_{\max }}<t_{A_{\max }}\) which holds in Table 5 except for data sets 8 and 10 from Bergamaschi et al. (2021). These values have been indicated in bold in Table 5.
We plot the data of each reference data set, along with the full simulated model (4) and each of the asymptotic approximations, in Fig. 3 for antibody data and Fig. 3 for IFN-\(\gamma \) and interleukin data. As with Figs. 1 and 2, we alternate gray–white backgrounds to showcase the different timescales. We plot the asymptotic solutions in each of the regions they are valid, but only include the time regions up to the terminal data point in each study. Based on the discussion surrounding Fig. 2c about practical asymptotic consistency, we extend the asymptotic solutions for \(t_2\) and \(t_4\) into the \(t_3\) regime because the practical validity can be different than what is theoretically predicted. In Fig. 3c, e, we do not include the \(t_4\) asymptotic solution in the \(t_3\) region because it is outside the plotting window. In Figs. 3 and 4 we also plot three vertical dashed lines which correspond to the maximal times (in chronological order) for each of the concentrations I, B, and A given in Table 5. If less than three lines are present it means that the predicted optima occur outside the time window where data was available.
Comparison of the full model (4), the analytic approximations described in Sect. 3 and data from the cited reference for IgG antibody A. Parameters and initial values are listed in Table 2 and Table 3 respectively (with corresponding number entry). The alternating patches are the timescale windows in Table 4. The vertical dashed lines are the times for the maximal concentrations in interleukin (pink), plasma B-cells (orange), and antibody (teal) from Table 5 ordered chronologically. We note the scales on the axes are different for some sub figures. Continued from previous page. Comparison of the full model (4), the analytic approximations described in Sect. 3 and data from the cited reference for IgG antibody A. Parameters and initial values are listed in Tables 2 and 3 respectively (with corresponding number entry). The alternating patches are the timescale windows in Table 4. The vertical dashed lines are the times for the maximal concentrations in interleukin (pink), plasma B-cells (orange), and antibody (teal) from Table 5 ordered chronologically. We note the scales on the axes are different for some sub figures
Comparison of the full model (4), the analytic approximations described in Sect. 3 and data from the cited reference for IFN-\(\gamma \) (F) and interleukin (I). Parameters and initial values are listed in Tables 2 and 3 respectively (with corresponding number entry). The alternating patches are the timescale windows in Table 4. The vertical dashed lines are the times for the maximal concentrations in interleukin, plasma B-cells, and antibody from Table 5 ordered chronologically. We note the scales on the axes are different for some sub figures
4.3 Multiple doses
We note that the original model (1) as posed does not allow for multiple doses which is seemingly problematic as all patient data used in Sect. 4.2 does include a second dose with second dosage times given in Table 3. Mathematically, it is straight-forward to account for additional doses in the model by including impulse terms to the LNP compartment of the model. However, when we compare the model with one and two doses to the data we seem to get a paradoxical conclusion that the one-dose model fits better despite the patient having received two doses (see Fig. 5 for an example). One possible explanation is that the parameters fit in Korosec et al. (2022) have accounted for a second dose through fitting. Furthermore, the second dosage time is in the \(t_4\) timescale where the peak of antibody occurs, thus creating a potential resolution issue. However, since the second dose does not affect the immune response timescales, nor the general analysis completed here, we save any further discussion of multiple doses for future work.
5 Discussion
During the model formulation in Sect. 2.1 we identified parameters \(\lambda _\textrm{F}\), \(\lambda _\textrm{C}\), and \(\alpha _\textrm{L}\) as likely being unidentifiable due to very small pre-factors and hence neglected them in the reduced model (6). Table 2 confirms this hypothesis as each of these parameters are very small based on fit data. We further set each of the saturation parameters to zero under the notion that we would not be over-saturated in plasma B-cells and interleukin. These parameters are indeed generally small in Table 2 justifying the assumption.
A further assumption in the non-dimensional model (4) was that the parameters are \(\mathcal {O}(1)\) or smaller so that the model is sensibly scaled. We note that this is mostly true in Table 2 with exceptions to this noted in bold. Data set 14 from Bergamaschi et al. (2021) has an anomalous value for \(\alpha _\textrm{B}\). From Fig. 4f we observe that most of the data is concentrated around the peak with some later data corresponding to the second dose time. Since \(\alpha _\textrm{B}\) is a decay parameter it will have the most significant contribution on interleukin after the peak value. Furthermore, from Sect. 3.4 the quasi-steady interleukin concentration (37) depends linearly on \(\alpha _\textrm{B}\), however there is very little data in this region to inform the parameter. Therefore, the value of \(\alpha _\textrm{B}\) in data set 14 may be a consequence of poor data fidelity.
The more interesting anomalous parameters are those for \(\lambda _\textrm{B}\) (data sets 11, 12, and 13) and \(\kappa _\textrm{I}\) (data sets 12 and 13) as these all belong to data collected by Widge et al. (2021) for the mRNA-1273 vaccine. The data in this paper measured antibody response for various ages. Data set 11 is for ages 18–55, data set 12 ages 56–70, and data set 13 ages 70 and over. We see a dramatic difference between those under and over the age of 55. While age effects on response were discussed by Korosec et al. (2022), the parameter contrast was not as apparent in the dimensional form. \(\lambda _\textrm{B}\) and \(\kappa _\textrm{I}\) are associated with the autocatalytic production of plasma-B cells and ultimately antibodies. A higher value leads to more rapid production which due to the inhibiting effects on interleukin can also lead to quicker decay.
We note in Table 4 that the \(t_3\) timescale representing the onset of autocatalytic production starts several days later in the older-patient data sets of Widge et al. (2021) than the younger-patient data set, but the \(t_4\) timescale representing peak antibody starts a few days sooner. Therefore, overall, the time window over which antibody levels are high is much narrower for ages 56 and greater compared to those below this age. We also note that the youngest age group has \(\alpha _\textrm{B}\) as the slowest decay parameter compared to the two older data sets and thus does not have predicted interleukin cessation. There is no confirmation for this in the data set because interleukin data was not recorded for these cohorts.
Interestingly, the predicted maximal antibody concentrations from Table 5 are higher for the two older-age data sets of Widge et al. (2021) compared to the data set for those under 56 with the highest maximum concentration predicted in the 56–70 data set. However, the decay rates \(\alpha _\textrm{B}\) and \(\alpha _\textrm{T}\) also increase, as observed in Table 2 and therefore a high concentration will also experience rapid decay, potentially explaining this counter intuitive result and providing insight into the body’s immunosenescence. We find that \(\lambda _\textrm{B}\) and \(\lambda _\textrm{I}\) seem to be age-dependent. These parameters dictate how well the body transitions from vaccine priming (the adapted immunity response) to self-production of antibody precursors (the innate immune response) and therefore provide detail on the immunogenicity of the vaccine. Overall, the analysis indicates the existence of, and possible mechanism for, an age dependent vaccine and natural immune response. A reduced vaccine efficacy with age has been reported in studies such as by Menni et al. (2022).
The anomalous bold values in Table 2 also skew the mean values for the mRNA-1273 vaccine making it appear as if there is discernible difference between that and the BNT162b2 vaccine. However, if data sets 12 and 13 of Widge et al. (2021) are removed then the new mean for vaccine mRNA-1273 (Mean M* in Table 2) is much more comparable with the mean for BNT162b2, indicating that there is no appreciable difference between the vaccines in terms of immune response parameters. It may appear in Table 5 that there are discernible differences in peak antibody levels between the two vaccines as computing the means for the two vaccines shows a nearly 20 fold difference between the two. However, this comparison cannot be made because the antibody concentrations that are reported are in arbitrary units and may be different for each study. While it is omitted here, redoing the analysis on the maximal antibody concentration in dimensionless units shows approximately a two-fold difference which makes them more comparable.
The data sets of Suthar et al. (2022) separated antibody response by sex with data set 19 being male and data set 20 being female. We see, both from the parameters in Table 2 and timescales in table 4, that there is no discernible difference between sexes in the vaccine response.
Interestingly, aside from the two over-55 groups of Widge et al. (2021), none of the mRNA-1273 studies considered here have interleukin cessation, whereas all of the BNT162b2 studies do with the exception of those by Suthar et al. (2022). However, it is worth noting that the studies by Suthar et al. (2022) also have data at the latest time points of all of the studies considered. Overall, the late-time study of interleukin concentration in each of the mRNA-1273 and BNT162b2 vaccines would be an interesting exploration.
Non-anomalous \(\lambda _\textrm{B}\) values in Table 2 are almost all \(\mathcal {O}(1)\). The three smallest values are of order \(\mathcal {O}(10^{-2})\) and belong to IFN-\(\gamma \) data set 6 and interleukin data sets 8 and 10. It is not surprising that IFN-\(\gamma \) parameter fitting may not capture a representative value of \(\lambda _\textrm{B}\) as the profile entirely mimics that of the CD4\(^+\) T-cell population whose dynamics do not depend on \(\lambda _\textrm{B}\). As for the interleukin data sets, observing Fig. 4c, e, we note that the graphs indeed look strange. Unlike the other interleukin data, there is no early peak, likely due to influence from the later data when the second dose is administered. Figure 4d, f where \(\lambda _\textrm{B}\sim \mathcal {O}(1)\) do have early peaks in the model simulation. While we do not plot it here, changing the value of \(\lambda _\textrm{B}\) for data sets 8 and 10 to be of \(\mathcal {O}(1)\) indeed introduces a peak into the model consistent with the other data sets. Another indication that the fitting to data sets 8 and 10 is problematic is observed in Table 5 where for those particular data sets we have the anomalous result that \(t_{B_{\max }}<t_{I_{\max }}\) counter to the other data sets and overall model analysis. Overall, the scaling analysis allows us to interrogate parameters far from the mean and analyze their validity.
Unlike the parameter \(\lambda _\textrm{B}\), the values of \(\lambda _\textrm{I}\) are much smaller than \(\mathcal {O}(1)\) generally, except for the anomalous data sets 12 and 13 previously discussed. Even though the analysis assumed \(\lambda _\textrm{I}\sim \mathcal {O}(1)\) the fact that \(\lambda _\textrm{I}\ll 1\) does not impact the overall immune response, it just creates some additional complexity between timescales \(t_3\) and \(t_4\). Specifically, if \(\lambda _\textrm{B}\sim \mathcal {O}(1)\) and \(\lambda _\textrm{I}\ll 1\) then when the \(t_3\) timescale begins, the plasma B-cell concentration is very high but the interleukin concentration remains small because of the weak non-linear decay. As such, the plasma B-cell concentrations grow explosively during which a new timescale \(t_3^*\) will emerge. This timescale separates from \(t_3\) similar to the interleukin cessation scale, and the concentration of plasma B-cells will become large enough that the weak interleukin inhibition becomes dominant. Overall, the interleukin activator-inhibitor dynamics proceed sequentially from activation to inhibition rather than concurrently as was presented here. Since the overall behaviour is captured by the limit we have chosen here, we omit details of the more general case.
Many of the insights discussed on data fidelity and parameter estimates are due to the explicit analytic expressions obtained. These expressions also provide biological insight. For example, through (45) we understand the minimum time for interleukin to begin inhibiting antibody production and that it depends on two dimensionless groupings, \(\lambda _\textrm{B}\) and \(\lambda _\textrm{I}\). We discussed that the peak antibody response seems to highly correlate with age and the result of (45) provides a reduced set of parameters to investigate age dependence in. Our study also showed the delicate relationship that each of the decay parameters have to long-term immune behaviour. If B-cells decay quickest so that \(\alpha _\textrm{B}\) is the largest decay parameter then quasi-steady interleukin concentrations can persist for a long time. Otherwise, interleukin decays quickly. This means that interleukin concentration data at long time points after vaccination can provide insight into the whether the system is dominated by T-cell or B-cell decay.
5.1 Conclusions and future work
Overall, we have presented a model for LNP delivered mRNA vaccines originally derived in Korosec et al. (2022). Through non-dimensionalisation we were able to reduce the model and identify five distinct timescales where different parts of the immune response dominated. Comparing our model to data we were able to identify data sets with different immune response, namely those associated with age, and also identify parameter sets which significantly differ from the mean.
Identifying important timescales of the immune response dynamics provides a framework for clinical data collection. The timescales are determined from two parameters, the rate of vaccine conversion in cells, \(\mu _\textrm{LV}\), and the decay rate of IgG antibodies, \(\gamma _\textrm{A}\). If those can be measured independently then timescales can be estimated and clinicians have windows with which to collect data. This avoids unnecessary sampling and missing important dynamics. Furthermore, it increases the likelihood of capturing data related to peak concentrations which can lead to stronger parameter estimation and thus stronger model prediction. If estimates of \(\mu _\textrm{LV}\) and \(\gamma _\textrm{A}\) are not available because of novel or emerging diseases, we recommend a two-stage trial. The first will create coarse window estimates based on parameters from comparative diseases. Parameter estimation will be done on these windows to refine estimates of the timescales at which point the refined trials discussed above can be conducted.
Data availability
The datasets generated during and/or analysed during the current study are available in the GITHUB repository, https://github.com/ionmoles/mRNA-inhost.
Code availability
The code generated during the current study are available in the GITHUB repository, https://github.com/ionmoles/mRNA-inhost.
References
Abramowitz M, Stegun IA (1983) Handbook of mathematical functions with formulas, graphs, and mathematical tables, vol 55. National Bureau of Standards, Dover Publications
Andrews GE, Askey R, Roy R et al (1999) Special functions, vol 71. Cambridge University Press
Bahl K, Senn JJ, Yuzhakov O et al (2017) Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther 25(6):1316–1327. https://doi.org/10.1016/j.ymthe.2017.03.035
Bergamaschi C, Terpos E, Rosati M et al (2021) Systemic IL-15, IFN-\(\gamma \), and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep 36(6):109,504. https://doi.org/10.1016/j.celrep.2021.109504
Betti M, Bragazzi NL, Heffernan JM et al (2021) Integrated vaccination and non-pharmaceutical interventions based strategies in Ontario, Canada, as a case study: a mathematical modelling study. J R Soc Interface 18(180):20210,009. https://doi.org/10.1098/rsif.2021.0009
Childs L, Dick DW, Feng Z et al (2022) Modeling waning and boosting of COVID-19 in Canada with vaccination. Epidemics 39(100):583. https://doi.org/10.1016/j.epidem.2022.100583
Dick DW, Childs L, Feng Z et al (2021) COVID-19 seroprevalence in Canada modelling waning and boosting COVID-19 immunity in Canada a Canadian immunization research network study. Vaccines 10(1):17. https://doi.org/10.3390/vaccines10010017
Ehreth J (2003) The global value of vaccination. Vaccine 21(7–8):596–600. https://doi.org/10.1016/S0264-410X(02)00623-0
Fair KR, Karatayev VA, Anand M et al (2022) Estimating COVID-19 cases and deaths prevented by non-pharmaceutical interventions, and the impact of individual actions: a retrospective model-based analysis. Epidemics 39(100):557. https://doi.org/10.1016/j.epidem.2022.100557
Farhang-Sardroodi S, Korosec CS, Gholami S et al (2021) Analysis of host immunological response of adenovirus-based COVID-19 vaccines. Vaccines 9(8):861. https://doi.org/10.3390/vaccines9080861
Foon KA, Sherwin SA, Abrams PG et al (1985) A phase I trial of recombinant gamma interferon in patients with cancer. Cancer Immunol Immunother 20(3):193–197. https://doi.org/10.1007/BF00205575
Gholami S, Korosec CS, Farhang-Sardroodi S et al (2023) A mathematical model of protein subunits COVID-19 vaccines. Math Biosci 358:108970
Goel RR, Apostolidis SA, Painter MM et al (2021) Distinct antibody and memory B cell responses in SARS-CoV-2 Naïve and recovered individuals after mRNA vaccination. Sci Immunol 6(58):eabi6950. https://doi.org/10.1126/sciimmunol.abi6950
Gonias SL, Pizzo SV, Hoffman M (1988) Clearance and distribution of recombinant murine \(\gamma \)-interferon in mice. Cancer Res 48(8):2021–2024
Heffernan J, Keeling M (2009) Implications of vaccination and waning immunity. Proc R Soc B Biol Sci 276(1664):2071–2080. https://doi.org/10.1098/rspb.2009.0057
Hernandez-Vargas EA, Velasco-Hernandez JX (2020) In-host mathematical modelling of COVID-19 in humans. Annu Rev Control 50:448–456. https://doi.org/10.1016/j.arcontrol.2020.09.006
Herz A, Bonhoeffer S, Anderson RM et al (1996) Viral dynamics in vivo: limitations on estimates of intracellular delay and virus decay. Proc Natl Acad Sci 93(14):7247–7251. https://doi.org/10.1073/pnas.93.14.7247
Hogan AB, Winskill P, Watson OJ et al (2021) Within-country age-based prioritisation, global allocation, and public health impact of a vaccine against SARS-CoV-2: a mathematical modelling analysis. Vaccine 39(22):2995–3006. https://doi.org/10.1016/j.vaccine.2021.04.002
Ke R, Martinez PP, Smith RL et al (2022) Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Nat Microbiol 7(5):640–652. https://doi.org/10.1038/s41564-022-01105-z
Kim KS, Ejima K, Iwanami S et al (2021) A quantitative model used to compare within-host SARS-CoV-2, MERS-CoV, and SARS-CoV dynamics provides insights into the pathogenesis and treatment of SARS-CoV-2. PLoS Biol 19(3):e3001,128. https://doi.org/10.1371/journal.pbio.3001128
Korosec CS, Farhang-Sardroodi S, Dick DW et al (2022) Long-term durability of immune responses to the BNT162b2 and mRNA-1273 vaccines based on dosage, age and sex. Sci Rep 12(1):21,232
Korosec CS, Betti MI, Dick DW et al (2023) Multiple cohort study of hospitalized SARS-CoV-2 in-host infection dynamics: parameter estimates, identifiability, sensitivity and the eclipse phase profile. J Theor Biol. https://doi.org/10.1016/j.jtbi.2023.111449
Kummer EE (1837) De integralibus quibusdam definitis et seriebus infinitis. J für die Reine und Angewandte Mathematik (Crelles J) 1837(17):228–242
Li Q, Tang B, Bragazzi NL et al (2020) Modeling the impact of mass influenza vaccination and public health interventions on COVID-19 epidemics with limited detection capability. Math Biosci 325(108):378. https://doi.org/10.1016/j.mbs.2020.108378
Lin J, Law R, Korosec CS et al (2022) Longitudinal assessment of SARS-CoV-2 specific T cell cytokine-producing responses for 1 year reveals persistence of multi-cytokine proliferative responses, with greater immunity associated with disease severity. J Virol. https://doi.org/10.1128/jvi.00509-22
Lindgren G, Ols S, Liang F et al (2017) Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ t follicular helper cells. Front Immunol. https://doi.org/10.3389/fimmu.2017.01539
Lozano-Ojalvo D, Camara C, Lopez-Granados E et al (2021) Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in Naive and COVID-19 recovered individuals. Cell Rep 36(8):109,570. https://doi.org/10.1016/j.celrep.2021.109570
Lutz J, Lazzaro S, Habbeddine M et al (2017) Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. npj Vaccines. https://doi.org/10.1038/s41541-017-0032-6
Menni C, May A, Polidori L et al (2022) COVID-19 vaccine waning and effectiveness and side-effects of boosters: a prospective community study from the ZOE COVID study. Lancet Infect Dis. https://doi.org/10.1016/S1473-3099(22)00146-3
Moore S, Hill EM, Tildesley MJ et al (2021) Vaccination and non-pharmaceutical interventions for COVID-19: a mathematical modelling study. Lancet Infect Dis 21(6):793–802. https://doi.org/10.1016/s1473-3099(21)00143-2
Moss R, Wood J, Brown D, et al (2020) Modelling the impact of COVID-19 in Australia to inform transmission reducing measures and health system preparedness https://doi.org/10.1101/2020.04.07.20056184
Moyles IR, Heffernan JM, Kong JD (2021) Cost and social distancing dynamics in a mathematical model of COVID-19 with application to Ontario, Canada. R Soc Open Sci 8(2):201,770. https://doi.org/10.1098/rsos.201770
Ndeupen S, Qin Z, Jacobsen S et al (2021) The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. Iscience 24(12):103,479. https://doi.org/10.1016/j.isci.2021.103479
Néant N, Lingas G, Hingrat QL et al (2021) Modeling SARS-CoV-2 viral kinetics and association with mortality in hospitalized patients from the French COVID cohort. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.2017962118
Pardi N, Tuyishime S, Muramatsu H et al (2015) Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release 217:345–351. https://doi.org/10.1016/j.jconrel.2015.08.007
Pardi N, Hogan MJ, Naradikian MS et al (2018) Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med 215(6):1571–1588. https://doi.org/10.1084/jem.20171450
Pardi N, Hogan MJ, Porter FW et al (2018) mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov 17(4):261–279. https://doi.org/10.1038/nrd.2017.243
Perelson AS (2002) Modelling viral and immune system dynamics. Nat Rev Immunol 2(1):28–36. https://doi.org/10.1038/nri700
Perelson AS, Ke R (2021) Mechanistic modeling of SARS-CoV-2 and other infectious diseases and the effects of therapeutics. Clin Pharmacol Ther 109(4):829–840. https://doi.org/10.1002/cpt.2160
Polyanin AD, Zaitsev VF (2017) Handbook of ordinary differential equations: exact solutions, methods, and problems. Chapman and Hall/CRC
Psimadas D, Georgoulias P, Valotassiou V et al (2012) Molecular nanomedicine towards cancer: \(^111\)In-labeled nanoparticles. J Pharm Sci 101(7):2271–2280. https://doi.org/10.1002/jps.23146
Rodrigues C, Plotkin SA (2020) Impact of vaccines; health, economic and social perspectives. Front Microbiol 11:1526. https://doi.org/10.3389/fmicb.2020.01526
Sadria M, Layton AT (2021) Modeling within-host SARS-CoV-2 infection dynamics and potential treatments. Viruses 13(6):1141. https://doi.org/10.3390/v13061141
Slater LJ (1960) Confluent hypergeometric functions. Cambridge University Press
Smirnova A, DeCamp L, Chowell G (2021) Mathematical and statistical analysis of doubling times to investigate the early spread of epidemics: application to the COVID-19 pandemic. Mathematics 9(6):625. https://doi.org/10.3390/math9060625
Stankov MV, Cossmann A, Bonifacius A et al (2021) Humoral and cellular immune responses against severe acute respiratory syndrome coronavirus 2 variants and human coronaviruses after single BNT162b2 vaccination. Clin Infect Dis 73(11):2000–2008. https://doi.org/10.1093/cid/ciab555
Suthar MS, Arunachalam PS, Hu M et al (2022) Durability of immune responses to the BNT162b2 mRNA vaccine. Med 3(1):25–27. https://doi.org/10.1016/j.medj.2021.12.005
Tang B, Bragazzi NL, Li Q et al (2020) An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov). Infect Dis Model 5:248–255. https://doi.org/10.1016/j.idm.2020.02.001
Tricomi F (1947) Sulle funzioni ipergeometriche confluenti. Annali di Matematica Pura ed Applicata 26(1):141–175
Vignals C, Dick DW, Thiébaut R et al (2021) Barrier gesture relaxation during vaccination campaign in France: modelling impact of waning immunity. COVID 1(2):472–488. https://doi.org/10.3390/covid1020041
Wang Z, Schmidt F, Weisblum Y et al (2021) mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592(7855):616–622. https://doi.org/10.1038/s41586-021-03324-6
Wells CR, Townsend JP, Pandey A et al (2021) Optimal COVID-19 quarantine and testing strategies. Nat Commun. https://doi.org/10.1038/s41467-020-20742-8
Widge AT, Rouphael NG, Jackson LA et al (2021) Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N Engl J Med 384(1):80–82. https://doi.org/10.1056/NEJMc2032195
Yuan P, Aruffo E, Gatov E et al (2022) School and community reopening during the COVID-19 pandemic: a mathematical modelling study. R Soc Open Sci 9(211):883. https://doi.org/10.1098/rsos.211883
Yuan P, Li J, Aruffo E et al (2022) Efficacy of a “stay-at-home’’ policy on SARS-CoV-2 transmission in Toronto, Canada: a mathematical modelling study. CMAJ Open 10(2):E367–E378. https://doi.org/10.9778/cmajo.20200242
Zhang C, Maruggi G, Shan H et al (2019) Advances in mRNA vaccines for infectious diseases. Front Immunol. https://doi.org/10.3389/fimmu.2019.00594
Acknowledgements
The authors are grateful to James Watmough, Lindi Wahl, David Dick, and Samaneh Gholami for fruitful discussions during the completion of this manuscript.
Funding
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grants (IRM, JMH) an NSERC Emerging and Infectious Disease Modelling Initiative (IRM, JMH), and the NSERC postdoctoral fellowship program (CSK).
Author information
Authors and Affiliations
Contributions
All authors devised the study and performed the literature review. IRM did the primary analysis and coding. All authors edited the manuscript and provided feedback.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A Plasma B-cell concentration in the \({t_{{3}}}\) Timescale
The autocatatlyic problem is given by (20) in Sect. 3.3 with timescale \(t=\epsilon ^{-1/3}{t_{{3}}}\). We pose an expansion \({B_{{3}}}\sim {B_{{3}}}_0+\epsilon ^{1/3}{B_{{3}}}_1\) where the leading order problem is given by (25) in Sect. 3.3,
As this is a non-linear Riccati equation we can simplify using the transformation \({B_{{3}}}_0=\tfrac{u_{{t_{{3}}}}}{\lambda _\textrm{I}u}\) which allows us to write (A1) as
where prime indicates differentiation with respect to \({t_{{3}}}\). Further transforming \(u={t_{{3}}}w(z)\) with \(z=\tfrac{(\lambda _\textrm{I}+\lambda _\textrm{B}){t_{{3}}}^3}{6}\) yields,
which is Kummer’s differential equation and has as solutions Kummer’s functions \(\textrm{M}(x,y,z)\) and \(\textrm{U}(x,y,z)\) (see Kummer 1837; Tricomi 1947; Slater 1960; Polyanin and Zaitsev 2017),
for arbitrary constants \(C_M\) and \(C_U\). Therefore, \({B_{{3}}}_0\) has solution,
which from (15a) must satisfy \({B_{{3}}}_0\sim \frac{{t_{{3}}}^2}{2}=\frac{6^{2/3}}{2(\lambda _\textrm{I}+\lambda _\textrm{B})^{2/3}}z^{2/3}\) for \(z\ll 1\). Substituting (A4) into (A5) and expanding for \(z\ll 1\) yields
to leading order where \(\Gamma (z)\) is the usual Gamma function (see for example Polyanin and Zaitsev 2017). (A6) does not match the required form of \({B_{{3}}}\) for \(z\ll 1\) and therefore we must choose \(C_U\) so that this term is zero,
Returning to the expansion (A6) the next order term is
where we have used the definition of b from (A3). Substituting this into (A5) we get that to leading order \({B_{{3}}}_0\) satisfies,
which satisfies the correct matching condition for all \(C_M\). Therefore, without loss of generality we can take \(C_M\)=1. Overall then, the solution for \({B_{{3}}}_0\) satisfies
1.1 No loss of asymptotic consistency
The next order problem of (20) is
where once again multiplying (A11a) by \(\lambda _\textrm{I}\) and (A11b) by \(\lambda _\textrm{B}\) and adding produces a reduced equation
Substituting this into (A11a) and recognizing from (22) that \({B_{{3}}}_0=\frac{\textrm{d}^{}{A_{{3}}}_0}{\textrm{d} {t_{{3}}}^{}}\) yields an equation for \({B_{{3}}}_1\),
which is linear and has solution,
We already know from the solution for the CD4\(^+\) T-cells in the \({t_{{3}}}\) timescale given by (19) that an important timescale emerges when \({t_{{3}}}\sim \mathcal {O}(\epsilon ^{-2/3})\) (\(t\sim \epsilon ^{-1}\)). However, in Sect. 3.2 the solution for B, A, and I broke down sooner thus introducing the \({t_{{3}}}\) timescale. It is therefore important to investigate any additional timescales that may emerge between \(t\sim \mathcal {O}(1)\) and \(t\sim \mathcal {O}(\epsilon ^{-1})\). To do so we investigate \({B_{{3}}}_0\) and \({B_{{3}}}_1\) for \({t_{{3}}}\gg 1\).
The asymptotic expansions of the Kummer functions for large argument are (see for example Abramowitz and Stegun 1983; Andrews et al. 1999; Polyanin and Zaitsev 2017),
and so from (26) for \({t_{{3}}}\gg 1\) we have that
and also from (22) that,
With these asymptotic expansions then, from (A14), we have that,
and also that
G(t) can be written in terms of an incomplete Gamma function (see Polyanin and Zaitsev 2017) and thus the first term in (A19) vanishes for \({t_{{3}}}\gg 1\). Thus, overall we have that
and \({B_{{3}}}\) does not lose asymptotic consistency at least up to \(\mathcal {O}(\epsilon ^{1/3})\) since \({t_{{3}}}<{t_{{3}}}^2\) for \({t_{{3}}}\gg 1\). Similarly from (21b) and (A11b) we have the expressions for interleukin under \({t_{{3}}}\gg 1\) satisfy \({I_{{3}}}_0={I_{{3}}}_1=0\) and therefore
Thus, interleukin also does not lose asymptotic consistency up to the order considered. Finally from (8), since A is bounded from above by the integral of B then since B does not lose asymptotic consistency then neither does A.
Therefore, there are no additional intermediate timescales between \(t\sim \mathcal {O}(\epsilon ^{-1/3})\) and \(t\sim \mathcal {O}(\epsilon ^{-1})\) for new dynamics to occur.
Appendix B Equal decay parameters
Here we consider the case when the parameters \(\alpha _i\) are not unique. For simplicity we will consider the case when they are all equal to each other, i.e. \(\alpha _i=\alpha \) recognizing that sub cases mixing equality and inequality will result in a combination of this analysis and that presented in Sect. 3. The reduced model from (6) is
subject to initial conditions \(V(0)=T(0)=B(0)=0\), \(L(0)=1\), \(A(0)=\mathcal {A}\), and \(I(0)=\mathcal {I}\). We note that we have removed the equation for F as it is unchanged from Sects. 3.1 and 3.2 where \(F\sim T\). We have also removed the equation for C since when \(\alpha _\textrm{T}=\alpha _\textrm{C}=\alpha \) then \(C=T\) As with (6) we can solve directly for L, V, and T,
and if we knew B then the antibodies would still be given by (8). Thus we need only to solve for B and I in (B22) which are analyzed through each of the timescale regimes in Sect. 3.
We begin following Sect. 3.2 where \(t\sim \mathcal {O}(1)\). The original expansion of T when \(\alpha _\textrm{V}\ne \alpha _\textrm{T}\) given by (13) is not impacted by the singularity structure of \(\alpha _\textrm{V}=\alpha _\textrm{T}\). As such the solution when the parameters are equal is the same as that in Sect. 3.2 when \(\alpha _\textrm{V}=\alpha _\textrm{T}=\alpha \). Since the loss of asymptotic consistency was not related to the \(\alpha _i\) parameters we still have breakdown when \(t\sim \mathcal {O}(\epsilon ^{-1/3})\), and for the \({t_{{3}}}\) timescale in Sect. 3.3, once again the singularity structure does not play a role when expanding T. Furthermore, since we only considered the leading order behaviour which did not depend on \(\alpha _\textrm{V}\) or \(\alpha _\textrm{T}\) then the solutions for B and I are the same to leading order, (26) and (24) respectively.
The \(t_4\) timescale of Sect. 3.4, where peak and decay of antibody occurs, is where the \(\alpha _i\) parameters have a dominant role and the singularity structure appears in (27) for example. When the parameters are the same then substituting \(t=\epsilon ^{-1}t_4\) into (B23c) and expanding for \(\epsilon \ll 1\) yields
From here, the setup follows that of Sect. 3.4 with the equations to solve given by (28) leading to interleukin quasi-steady state (29) and differential equation for the plasma B-cells given by (30) with the change in all three that \(T_{4_0}\) is replaced by \(\hat{T}_{4_0}\). The plasma B-cell solution becomes
which we observe still satisfies the required (32) for \(t_4\ll 1\). From (33) we obtain the antibody concentration
which itself has a singularity issue if \(\alpha =1\). In this particular case the solution reduces to
When all of the parameters are equal then \(\min (\alpha _\textrm{V},\alpha _\textrm{T},\alpha _\textrm{B})=\min (\alpha _\textrm{V},\alpha _\textrm{T})=\alpha \) and so the interleukin cessation timescale described in Sect. 3.5 should occur as the exponential decay rates for both CD4\(^+\) T-cells and plasma B-cells are identical. However, the identical decay rates also introduces algebraic terms and so the quasi-steady interleukin concentration (29) becomes
We observe the interesting result that when the parameters are equal the interleukin concentration still decays to zero, but with a power law decay rather than an exponential one.
The reduction of the solution in the \(t_4\) timescale allows an easy analysis of the maximal plasma B-cell concentration where, from (B25), we conclude that the maximal concentration of plasma B-cells occurs when \(t_4=2\alpha ^{-1}\) with maximal B-cell concentration \(B_{4_0}=2\alpha ^{-2}\lambda _\textrm{I}^{-1}(\lambda _\textrm{B}+\lambda _\textrm{I})\text {e}^{-2}\). For general \(\alpha \) the maximal concentration of antibody does not have a clean analytic form because of the mixture of polynomial and exponential terms in (B26). However, when \(\alpha =1\) then the antibody concentration is given by (B27) and has a single exponential term. Upon differentiating, the maximum concentration occurs at \(t_4=3\) for all \(\lambda _\textrm{B}\) and \(\lambda _\textrm{I}\) with maximal concentration given by \(A_{4_0}=9/2\lambda _\textrm{I}^{-1}(\lambda _\textrm{B}+\lambda _\textrm{I})\text {e}^{-3}\). The independence of the maximal concentration time on \(\lambda _\textrm{B}\) and \(\lambda _\textrm{I}\) is discussed in Sect. 4.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Moyles, I.R., Korosec, C.S. & Heffernan, J.M. Determination of significant immunological timescales from mRNA-LNP-based vaccines in humans. J. Math. Biol. 86, 86 (2023). https://doi.org/10.1007/s00285-023-01919-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00285-023-01919-3