Abstract
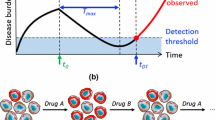
In previous work, we focused on the optimal therapeutic strategy with a pair of drugs which are collaterally sensitive to each other, that is, a situation in which evolution of resistance to one drug induces sensitivity to the other, and vice versa. Yoona (Bull Math Biol 8:1–34,Yoon et al. 2018) Here, we have extended this exploration to the optimal strategy with a collaterally sensitive drug sequence of an arbitrary length, N. To explore this, we have developed a dynamical model of sequential drug therapies with N drugs. In this model, tumor cells are classified as one of N subpopulations represented as \(\{R_i|i=1,2,...,N\}\). Each subpopulation, \(R_i\), is resistant to ‘\(Drug\ i\)’ and each subpopulation, \(R_{i-1}\) (or \(R_N\), if \(i=1\)), is sensitive to it, so that \(R_i\) increases under ‘\(Drug\ i\)’ as it is resistant to it, and after drug-switching, decreases under ‘\(Drug\ i+1\)’ as it is sensitive to that drug(s). Similar to our previous work examining optimal therapy with two drugs, we found that there is an initial period of time in which the tumor is ‘shaped’ into a specific makeup of each subpopulation, at which time all the drugs are equally effective (\(\mathcal {R}^*\)). After this shaping period, all the drugs are quickly switched with duration relative to their efficacy in order to maintain each subpopulation, consistent with the ideas underlying adaptive therapy. West(Canver Res 80(7):578–589Gatenby et al. 2009) and Gatenby (Cancer Res 67(11):4894–4903West et al. 2020). Additionally, we have developed methodologies to administer the optimal regimen under clinical or experimental situations in which no drug parameters and limited information of trackable populations data (all the subpopulations or only total population) are known. The therapy simulation based on these methodologies showed consistency with the theoretical effect of optimal therapy .









Similar content being viewed by others
References
Altrock PM, Liu LL, Michor F (2015) The mathematics of cancer: integrating quantitative models. Nature Rev Cancer 15(12):730–745
Archetti M, Ferraro DA, Christofori G (2015) Heterogeneity for igf-ii production maintained by public goods dynamics in neuroendocrine pancreatic cancer. Proc Natl Acad Sci 112(6):1833–1838
Basanta D, Simon M, Hatzikirou H, Deutsch A (2008) Evolutionary game theory elucidates the role of glycolysis in glioma progression and invasion. Cell Prolif 41(6):980–987
Brady R, Enderling H (2019) Mathematical models of cancer: when to predict novel therapies, and when not to. Bull Math Biol 81(10):3722–3731
Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL, Zhou L et al (2017) Early detection of molecular residual disease in localized lung cancer by circulating tumor dna profiling. Cancer Discovery 7(12):1394–1403
Cho H, Levy D (2018) Modeling continuous levels of resistance to multidrug therapy in cancer. Appl Math Modell 64:733–751
Cho H, Ayers K, de Pills L, Kuo Y-H, Park J, Radunskaya A, Rockne RC (2018) Modelling acute myeloid leukaemia in a continuum of differentiation states. Lett Biomath 5(sup1):S69–S98
Dhawan A, Nichol D, Kinose F, Abazeed ME, Marusyk A, Haura EB, Scott JG (2017) Collateral sensitivity networks reveal evolutionary instability and novel treatment strategies in alk mutated non-small cell lung cancer. Scientif Rep 7(1):1232
Dingle K, Camargo CQ, Louis AA (2018) Input-output maps are strongly biased towards simple outputs. Nature Commun 9(1):761
Enriquez-Navas PM, Kam Y, Das T, Hassan S, Silva A, Foroutan P, Ruiz E, Martinez G, Minton S, Gillies RJ et al (2016) Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci Trans Med 8(327):24–34
Gatenby RA, Silva AS, Gillies RJ, Frieden BR (2009) Adaptive therapy. Cancer Res 69(11):4894–4903
Gluzman M, Scott JG, Vladimirsky A (2020) Optimizing adaptive cancer therapy: dynamic programming and evolutionary game theory. Proc R Soc B 287(1925):20192454
Goldie J, Coldman A (1983) Quantitative model for multiple levels of drug resistance in clinical tumors. Cancer Treatment Rep 67(10):923–931
Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL (2001) Clinical resistance to sti-571 cancer therapy caused by bcr-abl gene mutation or amplification. Science 293(5531):876–880
Gottesman MM (2002) Mechanisms of cancer drug resistance. Ann Rev Med 53(1):615–627
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG (2013) Cancer drug resistance: an evolving paradigm. Nature Rev Cancer 13(10):714
Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, Sarkar S (2014) Drug resistance in cancer: an overview. Cancers 6(3):1769–1792
Iram S, Dolson E, Chiel J, Pelesko J, Krishnan N, Güngör Ö, Kuznets-Speck B, Deffner S, Ilker E, Scott JG et al. (2019) Controlling the speed and trajectory of evolution with counterdiabatic driving, bioRxiv, p. 867143
Kaznatcheev A, Vander Velde R, Scott JG, Basanta D (2017) Cancer treatment scheduling and dynamic heterogeneity in social dilemmas of tumour acidity and vasculature. Br J Cancer 116(6):785
Kaznatcheev A, Peacock J, Basanta D, Marusyk A, Scott JG (2019) Fibroblasts and alectinib switch the evolutionary games played by non-small cell lung cancer. Nature Ecol Evol 3(3):450
Khan KH, Cunningham D, Werner B, Vlachogiannis G, Spiteri I, Heide T, Mateos JF, Vatsiou A, Lampis A, Damavandi MD et al (2018) Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the prospect-c phase ii colorectal cancer clinical trial. Cancer Discovery 8(10):1270–1285
Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B (2005) Egfr mutation and resistance of non-small-cell lung cancer to gefitinib. New England J Med 352(8):786–792
Löscher W, Potschka H (2005) Drug resistance in brain diseases and the role of drug efflux transporters. Nature Rev Neurosci 6(8):591
Maltas J, Wood KB (2019) Pervasive and diverse collateral sensitivity profiles inform optimal strategies to limit antibiotic resistance. PLoS Biol 17(10):e33000515
Marusyk A, Polyak K (2010) Tumor heterogeneity: causes and consequences. Biochimica et Biophysica Acta (BBA)-Rev Cancer 1805(1):105–117
Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K (2014) Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514(7520):54–58
Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A (2014) Resistance to anti-egfr therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discovery 4(11):1269–1280
Nichol D, Jeavons P, Fletcher AG, Bonomo RA, Maini PK, Paul JL, Gatenby RA, Anderson AR, Scott JG (2015) Steering evolution with sequential therapy to prevent the emergence of bacterial antibiotic resistance. PLoS Comput Biol 11(9):e1004493
Nichol D, Rutter J, Bryant C, Hujer AM, Lek S, Adams MD, Jeavons P, Anderson AR, Bonomo RA, Scott JG (2019) Antibiotic collateral sensitivity is contingent on the repeatability of evolution. Nature Communi 10(1):334
Ogbunugafor CB, Wylie CS, Diakite I, Weinreich DM, Hartl DL (2016) Adaptive landscape by environment interactions dictate evolutionary dynamics in models of drug resistance. PLoS Comput Biol 12(1):874
Piddock LJ (2006) Multidrug-resistance efflux pumps? not just for resistance. Nature Rev Microbiol 4(8):629
Rejniak KA (2005) A single-cell approach in modeling the dynamics of tumor microregions. Math Biosci Eng 2(3):643–655
Rockne RC, Hawkins-Daarud A, Swanson KR, Sluka JP, Glazier JA, Macklin P, Hormuth DA II, Jarrett AM, Lima EA, Oden JT et al (2019) The 2019 mathematical oncology roadmap. Phys Biol 16(4):041005
Scott J, Marusyk A (2017) Somatic clonal evolution: a selection-centric perspective. Biochimica et Biophysica Acta (BBA)-Rev Cancer 1867(2):139–150
Scott JG, Fletcher AG, Anderson AR, Maini PK (2016) Spatial metrics of tumour vascular organisation predict radiation efficacy in a computational model. PLoS Comput Biol 12(1):e1004712
Scott JG, Sedor G, Scarborough JA, Kattan MW, Peacock J, Grass GD, Mellon EA, Thapa R, Schell M, Waller A et al (2021) Personalizing radiotherapy prescription dose using genomic markers of radiosensitivity and normal tissue toxicity in nsclc. J Thoracic Oncol 16(3):428–438
Tomasetti C, Levy D (2010) An elementary approach to modeling drug resistance in cancer. Math Biosci Eng: MBE 7(4):905
Trédan O, Galmarini CM, Patel K, Tannock IF (2007) Drug resistance and the solid tumor microenvironment. J Nat Cancer Instit 99(19):1441–1454
Vander Velde R, Yoon N, Marusyk V, Durmaz A, Dhawan A, Miroshnychenko D, Lozano-Peral D, Desai B, Balynska O, Poleszhuk J et al (2020) Resistance to targeted therapies as a multifactorial, gradual adaptation to inhibitor specific selective pressures. Nature Commun 11(1):1–13
West J, You L, Zhang J, Gatenby RA, Brown JS, Newton PK, Anderson AR (2020) Towards multidrug adaptive therapy. Cancer Res 80(7):1578–1589
Yoon N, Vander Velde R, Marusyk A, Scott JG (2018) Optimal therapy scheduling based on a pair of collaterally sensitive drugs. Bull Math Biol 8:1–34
Zhang J, Cunningham JJ, Brown JS, Gatenby RA (2017) Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nature Commun 8(1):1–9
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A Differential system of instantaneous drug switch
Definition
In addition to the definitions from Table 1, we defines further notations to facilitate the descriptions on proofs.
\(\mathcal {D}_i:=\mathcal {D}(i) \in \mathbb {R}^{N \times N}\), \(\mathcal {M}_i(t):=\left( m_{j,k}^i(t)\right) \in \mathbb {R}^{N \times N}\), \(\mathcal {L}_{i,\epsilon }:=\mathcal {M}_i(f_i \ \epsilon )\)
with
Also, we will use the following notation of product of an arbitrary sequence of matrices, \(\mathcal {A}_i\), \(\mathcal {A}_{i+1}\), \(\ldots \), \(\mathcal {A}_j\).
Proposition A.1
Under the therapy with \(Drug \ i\):
Proposition A.2
\(\left. {\mathcal {L}_{i,\epsilon }} \right| _{\epsilon =0} = I_N\) for all \(1 \le i \le N\).
Proposition A.3
\(\left. {\displaystyle \frac{d}{d\epsilon } \mathcal {L}_{i,\epsilon }} \right| _{\epsilon =0} = \displaystyle f_i \ \mathcal {D}_i\), for all \(1 \le i \le N\)
Lemma A.4
\(\displaystyle \frac{d}{d\epsilon } \left( \overset{\curvearrowleft }{\displaystyle \prod ^{i}_{k=1}} \mathcal {L}_{k,\epsilon } \right) =\sum _{k=1}^i f_k \ \mathcal {D}_k\) for all \(1 \le i \le N\)
Proof
\(\square \)
Lemma A.5
For any positive integer, n, and an any integer, i, in [1, N]
where \(\eta \) is some number in [0, 1) and \(0 \le f_k \le 1\) with \(\sum _{k=1}^N f_k=1\).
Proof
\(\square \)
Theorem A.6
If Drug1, Drug2, ..., DrugN, are prescribed in a cycle, and are switched instantaneously with relative duration of \(0 \le f_1\), \(f_2\), ..., \(f_N \le 1\) (where \(\sum _{k=1}^N f_k=1\)), respectively, \(\mathcal {R}\) obeys
Proof
For any time point \(t_0\), let us define \(\mathcal {R}_\epsilon (t)\) as a vector-valued function of \(R_1(t)\), \(R_2(t)\),..., and \(R_N(t)\) describing the cell population dynamics under a periodic therapy starting at \(t_0\) with \(Drug \ i\) assigned for \(t_0+\left( m+\sum _{k=1}^{i-1} f_i\right) \ \epsilon \le t < t_0+\left( m+\sum _{k=1}^i f_i\right) \epsilon \) where m is any non-negative integer and i is any integer in [1, N].
For any \(\Delta t>0\), there uniquely exist \(n\in \{0,1,2,...\}\) and \(\eta \in [0,1)\) such that \(\Delta t = \left( n + \sum _{k=1}^{i-1} f_k + \eta f_i\right) \ \epsilon \). Then, by Proposition A.1 and the definitions of \(\mathcal {L}_i\)s,
where \(\mathcal {R}(t_0)=\left( \begin{array}{cccc} R_1(t_0)&R_2(t_0)&\cdots&R_N(t_0) \end{array} \right) ^T\). And, \(\mathcal {R}_0(t)\) represents instantaneous drug switching.
Therefore,
\(\square \)
Theorem A.7
The population makeup at which all the drugs are equally effective is
where \(\{\mathcal {P}^1,\mathcal {P}^2,\ldots ,\mathcal {P}^N\}\) is linearly independent.
Proof
By the definition (4), the effect of \(Drug \ i\) is
Then, at a specific population makeup with balanced drug effects, denoted by \(\mathcal {R}^*\),
where k is a constant representing the level of balanced drug effect. Since \(\{\mathcal {P}^1,\mathcal {P}^2,\ldots ,\mathcal {P}^N\}\) is linearly independent, we can isolate \(\mathcal {R}^*\) with an inverse matrix.
Also, \(||\mathcal {R}^*||_1=1\), therefore
and
Corollary A.8
When the drugs are symmetric, the population makeup at which all the drugs are equally effective is
where \(\{\mathcal {P}^1,\mathcal {P}^2,\ldots ,\mathcal {P}^N\}\) is linearly independent.
Proof
Similar to the proof of Theorem A.7, there exists unique solution of \(\mathcal {R}^*\) and k satisfying
If we plug in \(\mathcal {R}^* = \displaystyle \frac{1}{N} \ \varvec{1}\) to the equation,
Therefore, proved.
Theorem A.9
\(\{\mathcal {P}^1,\mathcal {P}^2,\ldots ,\mathcal {P}^N\}\) is linearly dependent, if the drugs are symmetric (i.e., \(\{p_r^i,p_s^i,p_0^i\}=\{p_r,p_s,p_0\}\) for all i) and \(p_r+p_s+(N-2)p_0=0\).
Proof
It suffices to show that there exists a linear combination of \(\mathcal {P}^i\)s that equals to a zero vector. Let the general form of a linear combination be \(\sum _{i=1}^N a_i\mathcal {P}^i\), where \(a_i\)s are constants. If \(a_i=1\) for all i, \(\sum _{i=1}^N \mathcal {P}^i=(p_r+p_s+(N-2)p_0)\varvec{1}=0 \cdot \varvec{1}=0\). Therefore, \(\{\mathcal {P}^1,\mathcal {P}^2,\ldots ,\mathcal {P}^N\}\) is linearly dependent.
Theorem A.10
Under instantaneously switching of symmetric drug cycle, total population with equal drug duration \((f_1=f_2=...=f_N=1/N)\), TP, is changing exponentially with the growth/decay rate,
Proof
The derivative of total population, TPD, is the summation of the vector, \(\frac{d\mathcal {R}}{dt}\).
\(\square \)
Appendix B Shannon entropy of periodic sequences
Shannon entropy is a measure of diversity of a heterogeneous collection. If there are k different types of elements in the multiset, and if we represent the proportions of the types as \(f_1\), \(f_2\), \(\ldots \) \(f_k\) (\(\sum _{i=1}^k f_i=1\)), respectively, its Shannon entropy is
If the multiset is composed by one identical element, its Shannon entropy is 0. Otherwise, if the multiset is composed by all distinct k elements, its Shannon entropy is \(\log k\).
We used this concept of the entropy to explore the level of periodicity shown on sequences. For a sequence \(a=(a_i)_{i\in \mathbb {N}}\), we define an associated sequence \((h_i|a)_{i\in \mathbb {N}}\) such that \(h_i\) is the entropy of the first part of the sequence, \((a_j)_{j=1}^i\). With any arbitrary \((a_i)_{i\in \mathbb {N}}\), \(h_1=0\). If \((a_i)_{i\in \mathbb {N}}\) is entirely periodic with the periodicity k, \(h_k=h_{2k}=\cdots =h_{nk}=\cdots (:=h)\), and \(\lim _{k \rightarrow \infty }h_{i}=h\). Figure 10 describes \((h_j|a)_{i\in \mathbb {N}}\) of an example of periodic sequence \(a=(a_i)_{i\in \mathbb {N}}\).
Such idea of entropy evaluation was applied for all the stages (4 stages in total) of the drug sequences in the exercise of optimal therapy simulation shown on Fig. 6. (Figure 11) The first stage is comprised by only one type of drug, therefore the entropy stays 0. In each of other stages, the corresponding sequence of entropy is nonzero, and is stabilized to the entropy level of the entire sequences of the stage. Also, we can see the periodic oscillations as the sequence proceeds. This observation is not quantified, however we believe it is sufficient to say the drug sequence within each stage is almost" periodic. As expected, such pattern is not observed, when we randomly shuffle and rearrange the drug sequences within stages (Fig. 12a), or rearrange the drugs in the order of \(Drug\ 1\), \(Drug\ 2\), \(Drug\ 3\) and \(Drug\ 4\) (Fig. 12b).
Entropy sequences of the 4 stages of drug sequences of Fig. 6. The horizontal gray lines present the entropy values of the entire sequence of the 4 stages
Rights and permissions
About this article
Cite this article
Yoon, N., Krishnan, N. & Scott, J. Theoretical modeling of collaterally sensitive drug cycles: shaping heterogeneity to allow adaptive therapy. J. Math. Biol. 83, 47 (2021). https://doi.org/10.1007/s00285-021-01671-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00285-021-01671-6