Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) colonization increases the risk of infection. Response to decolonization treatment is highly variable and determinants for successful decolonization or failure of eradication treatment are largely unknown. Insight into genetic predictors of eradication failure is potentially useful in clinical practice. The aim of this study was to explore genetic characteristics that are associated with MRSA decolonization failure. This cohort study was performed in a tertiary care hospital in the Netherlands. Patients with ≥ 1 positive MRSA culture from any site and with available whole -genome sequencing data of the MRSA isolate between 2017 and 2022 were included. Lineages, resistance, and virulence factors were stratified by MRSA decolonization outcome. In total, 56 patients were included: 12/56 (21%) with treatment failure and 44/56 (79%) with successful decolonization (with or without preceding treatment). A significant association was found between ciprofloxacin-resistant lineages and failure of eradication (OR 4.20, 95%CI 1.11–15.96, P = 0.04). Furthermore, livestock-associated MRSA and the major community-associated MRSA lineages ST6-t304 and ST8-t008 were associated with successful eradication treatment or spontaneous clearance. In conclusion, this explorative study showed a higher eradication failure rate in complicated MRSA carriers with ciprofloxacin-resistant MRSA lineages, which are predominantly healthcare-associated. Further studies are warranted to confirm the higher eradication failure risk of ciprofloxacin-resistant lineages, and identify the underlying mechanisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a global health threat with high morbidity and mortality rates [1]. Colonization with MRSA leads to increased infection rates of up to 25% [2, 3]. The Netherlands has one of the lowest levels of endemic MRSA in the world [4]. This low prevalence is for a large part attributed to a successful ‘search and destroy’ policy aiming at MRSA carriage, that has been executed for over three decades [5]. This policy consists of screening and pre-emptive strict isolation of patients with increased risk of MRSA carriage when hospitalized and subsequent decolonization treatment when carriage is found. Response to decolonization treatment is highly variable; in some patients, eradication treatment fails despite multiple attempts, in others colonization is self-limiting without treatment [6, 7]. Spontaneous clearance or persistent carriership is driven by a complex host–pathogen interaction, which is largely unraveled. Furthermore, antimicrobial treatment (i.e., eradication therapy) adds to this complex interaction, and introduces pharmacodynamic and pharmacokinetic effects. In summary, patient characteristics, antibiotic regimen, and isolate characteristics are all considered to contribute to decolonization treatment outcomes [7,8,9].
Different MRSA clones have emerged throughout the world with a high variety in virulence factors [10]. The rapid developments in the field of genetic diagnostics, especially whole-genome sequencing (WGS), have expanded the knowledge of the complexity and heterogeneity of this pathogen. MRSA strains produce a broad range of virulence factors, such as toxins, immune evasion factors, and adhesion proteins [11]. These virulence determinants are mostly carried on mobile genetic elements (MGEs), such as pathogenicity islands, plasmids, or bacteriophages [3]. Furthermore, virulence determinants can vary between hospital-associated, community-associated, and livestock-associated (LA) MRSA strains [12].
WGS of MRSA strains has been deployed extensively for infection control purposes. It has proven to be of great value in the epidemiology and outbreak management of MRSA [13]. In addition, WGS allows for molecular characterization of isolates by identifying clinically relevant genetic determinants that can help to predict response to decolonization treatment. So far, microbial genomics is not yet broadly applied to identify determinants related to MRSA eradication treatment outcome [14]. As an example, the presence of Panton-Valentine leucocidin (PVL) genes and genes associated with mupirocin resistance were associated with successful eradication outcome [9, 15]. A recent study elaborated on genetic factors and carriage duration, and showed a potential role of bacteriophage-related chemotaxis inhibitory protein encoded by chp [8]. Insight into genetic predictors of eradication failure is potentially useful in clinical practice. Ultimately, differentiating between MRSA carriers that will benefit from an eradication treatment and carriers more prone to eradication failure may enable personalized medicine.
In this explorative pilot cohort study, we evaluated genomic characteristics that are associated with MRSA decolonization failure. This was established by linking WGS data of MRSA isolates to clinical patient characteristics.
Methods
This cohort study was conducted at the University Medical Center Groningen, a tertiary hospital in the Northern part of the Netherlands, between 2017 and 2022. The prevalence of MRSA carriage in the Netherlands during this time was < 1%. During these years, genetic analyses of first MRSA isolates (both from carriage and infection) had been performed in all index patients and most of the healthcare workers, for the purpose of surveillance and outbreak management. Genetic analysis was not performed in healthcare workers who were positive at their pre-employment screening nor in positive family contacts of index patients. All patients (both adults and children) and healthcare workers of whom WGS of an MRSA isolate was performed were retrospectively identified and were screened to meet the selection criteria. Healthcare workers will be also addressed as ‘patients’ from now on in this manuscript, since they were treated as patients for this matter. Inclusion criteria were ≥ 1 visit to the outpatient infectious diseases clinic because of MRSA carriage or infection, ≥ 1 positive MRSA culture from any site, and available WGS data of the MRSA isolate. Exclusion criterion was the absence of follow-up cultures. Only the first available MRSA isolate per patient was included in the analysis. The patients had been assessed by the outpatient clinicians using protocols based on the national MRSA eradication guideline [16]. This includes in case of an MRSA infection, adequately treating the infection first, and subsequently screen for persistent colonization.
Data Collection
Clinical data were extracted from the electronic patient files. This included demographics, complicated versus uncomplicated carriage, treatment regimen, duration of therapy, and follow-up cultures. MRSA culture results were extracted from the laboratory information system. This included initial and follow-up MRSA cultures, including minimal inhibitory concentrations (MICs) of antibiotics, phenotypic susceptibility results, and WGS results.
Microbiological Methods
Culturing using BHI broth with 2.5% saline and MRSAid chromagar (bioMérieux, Lyon, France), susceptibility determination by automated susceptibility testing by VITEK2 (bioMérieux, Lyon, France), and cefoxitin disk diffusion were performed according to the Dutch Society of Medical Microbiology guideline for laboratory detection of highly resistant microorganisms as part of routine diagnostic procedures [17]. MIC breakpoints and zone diameter breakpoints for resistance and intermediate sensitivity were based on EUCAST criteria [18]. The isolates were identified as S. aureus by matrix-assisted laser desorption/ionization–time of flight mass spectrometry (Bruker Daltonics, Billerica, US). First MRSA isolates per patient were genotypically confirmed by Xpert MRSA NxG based on the detection of the mecA or mecC targets (Cepheid, Sunnyvale, US).
A total DNA extraction for whole-genome sequencing was performed directly from colonies of the respective isolates using the Ultraclean Microbial DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, US) according to the manufacturer’s protocol. DNA concentrations were determined using a Qubit® 2.0 fluorometer and the dsDNA HS and/or BR assay kit (Life Technologies, Carlsbad, CA, US). Subsequently, DNA libraries were prepared using the Nextera XT v2 kit (Illumina, San Diego, CA, US) according to the manufacturer’s instructions. Short-read sequencing was performed with an Illumina MiSeq System generating paired-end reads of 250 bp. De novo assembly of paired-end reads was performed using CLC Genomics Workbench v12.0.1-v20.0.4 (QIAGEN, Hilden, Germany) after quality trimming (Qs ≥ 20) establishing a word size of 29.
Based on next generation sequencing data (ENA project number PRJEB59407), molecular typing was performed using Ridom Seqsphere + v8.3.1 (Ridom, Münster, Germany). Herewith multilocus sequence typing (MLST) ST type was derived and core genome multilocus sequence typing (cgMLST) was performed using a scheme including 1861 alleles [19]. Isolates with a maximum of 24 allelic differences were denominated the same complex type. Antibiotic resistance genes were identified by Resfinder v4.1 (Center for Genomic Epidemiology, Lingby, Denmark). A predefined set of virulence factors were identified using AlereMicroarray schemes in Ridom Seqsphere + v8.3.1 (Ridom, Münster, Germany) [20].
Definitions
Uncomplicated MRSA carriage was defined as having all of the following features: (i) the presence of MRSA exclusively located in the nose, (ii) no active infection with MRSA, (iii) in vitro susceptibility for mupirocin, (iv) the absence of active skin lesions, (v) the absence of foreign material that connects an internal body site with the outside (e.g., urine catheter, external fixation material), and (vi) no previously failure of decolonization treatment. All other carriage cases were considered complicated colonization. Uncomplicated carriage is advised to be treated with topical therapy (mupirocin topically applied to the nares, disinfecting shampoo) and hygienic measures. In cases of complicated MRSA carriage, additional systemic antimicrobial therapy with a combination of two antibiotic agents is recommended, according to the national guideline [16]. MRSA infection was defined as a positive culture send to the microbiology laboratory from an infected body site as indicated by the treating physician.
Successful decolonization was defined as three consecutive negative MRSA cultures from swabs taken from nose, throat, and perineum, with the cultures obtained at 1-week intervals, without antibiotic usage [16]. For analyses, patients were divided in two groups: patients with failure of eradication treatment (failure group) and patients with successful decolonization with or without preceding treatment (successful decolonization group).
Livestock-associated MRSA was defined based on the Spa-type. The Spa-types t011, t034, t108, t567, t571, t588, t753, t779, t898, t899, t943, t1184, t1197, t1254, t1255, t1451, t1456, t1457, t2123, t2287, t2329, t2330, t2383, t2582, t2748, t2971, t2974, t3013, t3014, t3053, t3146, and t3208 were considered to be associated with livestock [12]. All other Spa-types were considered to be not associated with livestock.
Statistical Analysis
Data are presented as percentages or proportions for categorical variables and as medians plus interquartile range (IQR) for continuous variables. Univariate analysis was performed using Fisher’s exact test. As this study has an explorative character, no adjustment for multiple testing was done.
Results
During the study period, 181 patients visited the MRSA outpatient clinic. WGS was performed in 56/181 (31%) patients and these were included in the study (Fig. 1). As shown in Fig. 1, there were 12 patients with treatment failure (i.e., one in the uncomplicated carriage group and eleven in the complicated carriage group). All other patients (44) were MRSA negative at the end of follow-up and were defined as successfully decolonized (three in the uncomplicated carriage group, eight with MRSA infection without subsequent carriage, ten with spontaneous decolonization and 23 with successful treatment of complicated carriage). Patient and treatment characteristics of these two groups are depicted in Table 1. In the failure group, one patient out of twelve (8%) had uncomplicated carriage and 11/12 (92%) patients had complicated carriage. The successful decolonization group existed of 33/44 (75%) patients with complicated carriage, 3/44 (7%) patients with uncomplicated carriage, and 8/44 (18%) patients with MRSA infection, without subsequent carriage. Twenty-six out of 44 (59%) patients successfully underwent eradication treatment, in 10/44 (23%) patients colonization resolved spontaneously and 8/44 (18%) were treated for an MRSA infection, without subsequent eradication treatment. Of all 34 patients who underwent eradication treatment for complicated MRSA carriage, 11/34 (32%) had treatment failure. No significant differences in treatment characteristics were found between patients with treatment success and treatment failure (Table 1).
Flow chart. Flow chart of inclusions. MRSA carriage was defined as complicated in 44/56 (79%) patients, of whom 34/44 (77%) received systemic antibiotics as eradication treatment. The other 10/44 (23%) patients with complicated carriage were spontaneously cleared of MRSA before start of the planned eradication treatment. In addition, 8/56 (14%) patients with MRSA infections did not have subsequent MRSA carriage, and 4/56 (7%) patients had uncomplicated carriage. The green numbers represent the successful decolonization group (with or without preceding treatment). The red numbers represent the treatment failure group
Lineages
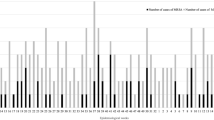
Among the 56 MRSA isolates, 24 different MLST types were represented. The most predominant MLST types were ST5 (8/56) and ST22 (8/56), followed by ST8 (5/56) and ST398 (5/56) (Fig. 2 and Table S1). The complex types were mostly unique, only seven complex types were represented twice (2615, 4940, 6749, 9359, 10,282, 17,413, 24,737). All isolates (n = 7) with livestock-associated Spa-types belonged to clonal complex 398. The non-livestock-associated MLST types ST1 (2/3), ST97 t2770 (2/2), ST6627 (1/1), and ST7119 (1/1) were more frequently or exclusively found in the failure group. In contrast, isolates of patients with successful decolonization predominantly belonged to community-associated lineages ST6-t304 (4/4), ST8-t008 (5/5), and the livestock-associated clonal cluster 398 (7/7) (Fig. 2).
Phylogenetic tree of MRSA isolates. Neighbor-joining tree from SeqSphere software based on curated schema where comparison of 1861 core genes of S. aureus was used. The study isolates from patients who failed on eradication treatment are presented in blue, and from patients with successful decolonization in pink. The corresponding isolate antibiotic susceptibility profiles are shown in supplementary table S1
Susceptibility and Resistance Genes
All MRSA isolates tested susceptible for the antibiotics used in the eradication treatments, and this was in line with the sequencing data that showed the absence of acquired resistance genes to these drugs (Table S2). Treatment failure was therefore not the result of resistance against the antibiotics used for the treatment. A significant association was found between ciprofloxacin resistance and failure of eradication (OR 4.20, 95%CI 1.11–15.96, P = 0.04) (Table 2). None of the patients had been treated with ciprofloxacin. The ciprofloxacin-resistant isolates belonged to ST5 (5), ST8 (2), ST22 (3), ST30 (1), ST97 (2), ST105 (1), ST398 (1), ST5544 (1), ST7119 (1), and ST8018 (1). In the ciprofloxacin-resistant isolates (n = 18), we detected one or more of the associated point-mutations S84L (10/18) in the gyrase GyrA, S80F (14/18) or S80Y (3/18) or E84G (2/18) or I45M (1/18) in the DNA topoisomerase IV GrlA, and P585S (1/18) in GrlB (Table S3). In the isolates of all patients with treatment failure, mutations associated with ciprofloxacin resistance were identified in 7/12 (58%) of the isolates, whereas in the isolates of patients with successful decolonization, these mutations were identified in 13/44 (30%) isolates (Fig. 3). Two isolates with the unique point mutation I45M in GrlA did not show increased MICs to ciprofloxacin. All seven persons with ciprofloxacin-resistant MRSA with failure to eradication treatment were either healthcare workers, or most likely had acquired the MRSA during hospitalization or after medical interventions. Rifampicin resistance-associated point-mutations were found in four isolates (I527L [3/4] and D471Y [1/4] in rpoB). While all four of these isolates had a rifampicin MIC ≤ 0.03, these isolates belonged to four patients with treatment failure (Table S3). No other associations were found between phenotypic antibiotic resistance or resistance genes and failure of eradication treatment (Table 3).
Ciprofloxacin MIC according to MRSA decolonization outcome and mutations associated with ciprofloxacin resistance. MIC range depicted by decolonization outcome and the presence of mutations (in GrlA, GrlB, or GyrA) associated with ciprofloxacin resistance. In the isolates of patients with treatment failure (red, n = 12), mutations were identified in 7/12 (58.3%) of the isolates, whereas in the isolates of patients with successful decolonization (green, n = 44), mutations were identified in 13/44 (29.5%) isolates. In isolates with high resistance (i.e., MIC > = 8mg/L) to ciprofloxacin (n = 9), multiple mutations were detected (Table S3), except for 1 isolate without mutations
Virulence Factors
An overview of the distribution of virulence genes among the patients with eradication failure and patients with successful decolonization is presented in Table 4. No associations were found between virulence genes and failure of eradication. Remarkably, PVL (lukF_PV and lukS_PV) was found more often in patients with successful decolonization compared to the patients with eradication failure, although non-significant (30% vs 17%, P = 0.48). The genes lukF_PV and lukS_PV and spIE were significantly associated with an MRSA infection (P < 0.05). The genes aur, hlgABC, icaACD, setB, setC, hlI, hlII, arcc, aroe, glpf, gmk, pta, tpi, yqil, isaB, lukX, lukY, and ebpS were present in all isolates and were therefore excluded from the analysis. The genes arc, edinABC, etABD, seb, sec, and sed were only sporadically present and were therefore excluded from the analysis as well.
Discussion
In this study, we explored associations between MRSA isolate characteristics, genetic determinants, and decolonization outcomes in a Dutch population of MRSA carriers in a tertiary hospital. We found an association of eradication failure with carriage of ciprofloxacin-resistant healthcare-associated lineages, whereas livestock-associated MRSA lineage ST398 and the majority of community-associated MRSA lineages ST6-t304 and ST8-t008 were associated with successful eradication treatment or spontaneous clearance.
The failure rate in eradication treatment of complex MRSA carriers was higher compared to previous reports in Dutch studies [5, 7]. Our study was conducted in the outpatient clinic of a tertiary hospital, with consequently a more than average representation of healthcare workers or patients with an extensive history of hospitalizations. Such patients mainly carry healthcare-associated MRSAs, that are adapted to survive under harsh nosocomial conditions and antibiotic exposure.
In our study, we found an association between ciprofloxacin resistance and failure in eradication treatment. Remarkably, none of the patients had been treated with ciprofloxacin. The ciprofloxacin-resistant MRSAs in our study belonged to various lineages, including five isolates of the healthcare-associated ST5 lineage with single amino acid substitution in GrlA S80F. The mutation in this healthcare-associated lineage, and its association with fluoroquinolone resistance and the presence of virulence genes as enterotoxins, β-hemolysin converting phage, and leucocidins has been described previously [21]. The resistance to fluoroquinolones is generally high in healthcare-associated MRSA [22]. Successful hospital-adapted ciprofloxacin-resistant lineages have emerged among several nosocomial species as E. coli, K. pneumoniae, vancomycin-resistant E. faecium, and MRSA. These lineages have acquired stable point-mutations in gyrase and/or topoisomerase IV enzymes [23]. It is unsure what drives this evolution, besides the exposure to fluoroquinolones.
Both tolerance and persistence have been reported in low-level ciprofloxacin-resistant E. coli, allowing to survive exposure to therapeutic concentrations of ciprofloxacin [24]. In tolerance, bacterial cells survive using a “hibernation mode,” in which the cell cycle and metabolism are temporarily stopped, preventing killing by antibiotics. In persistence, a bacterial subpopulation is able to survive antibiotic exposure [25]. Cross-tolerance to multi-drugs has been reported, but does not necessarily occur in all tolerant isolates and is dependent on antibiotic regimen and duration of exposure [26]. To the best of our knowledge, no studies have reported cross-tolerance in low-level ciprofloxacin-resistant S. aureus isolates to the antibiotic regimens in MRSA eradication used in this study. Therefore, the explanation for the association found in our study remains uncertain. Potentially, healthcare-associated MRSAs are more prone to failure of eradication treatment, and ciprofloxacin resistance may be a biomarker for these difficult-to-treat lineages.
The recent finding of association between chp and carriage duration was not found in our study [8]. Compared to the Danish study, our patient population had more healthcare-associated MRSA. Also, there is large heterogeneity in the Danish and Dutch MRSA treatment guidelines. The main difference is the more general use of two systemic antibiotics in the Netherlands, compared to sporadic systemic treatment in Denmark.
Two studies, in Denmark and Sweden, reported that PVL-positive isolates had a higher eradication success rate [15, 27]. We also found a higher (non-significant) rate of PVL-positive isolates in the successful eradication group, mainly belonging to the CA-MRSA linages ST30 and ST8-t008. However, associations do not necessarily reflect an etiologic cause, but can also reflect markers or confounders. We postulate that PVL is a marker of certain non-healthcare-associated MRSA lineages that are easier to eradicate, rather than a direct positive effect of the PVL toxin to eradication outcomes.
There are multiple factors of potential influence on MRSA eradication outcome. Carriers can reacquire MRSA isolates from contamination in their environment, or by positive household members. The eradication treatment of patients in this study was performed in a specialized outpatient clinic setting, following the Dutch eradication protocol [16]. Several measures are taken to prevent reacquisition, such as simultaneous treatment of positive household members and hygienic instructions. Isolate characteristics may also play a role in the risk of spread and reacquisition of MRSA. Hetem et al. showed that in a hospital setting, the transmission of livestock-associated MRSA was 4.4 times lower compared to non-livestock-associated MRSA isolates [12]. In general, MRSA isolates can be able to survive antibiotic exposure, despite having a MIC indicating susceptibility to the antibiotic agent. Our study showed that the antibiotic treatment failure is not explained by the common acquired resistance genes related to resistance, of which the presence or absence corresponded to the phenotypic susceptibility in all isolates. However, alternative survival mechanisms to antibiotic exposure, such as tolerance and persistence, are not detectable by measuring MICs. Other potential factors influencing MRSA eradication outcome, e.g., therapy incompliance and host genetics [28], were not assessed in our study.
There are some limitations of this study. It is a single-center study with a small sample size, a heterogeneous population, and a limited number of failed treatments. In addition, we did not always confirm that treatment failure was caused by the same clone, or acquisition of a different MRSA. However, given the very low prevalence of MRSA in the Netherlands, this would be highly unlikely. Furthermore, we did not correct for multiple testing. However, since it is an explorative study in a relatively undiscovered subject, we believe the results are still valid and useful in targeting future research. For this explorative purpose, we focused on pathogen factors and only added a limited number of host characteristics (i.e., sex, age, and complicated versus uncomplicated carriership). Other host factors—including host genetics—may influence the risk of treatment failure as well. Lastly, we investigated genes with a previously reported role in virulence. Future genome-wide association studies could perhaps identify signatures with novel genetic factors implicated in intracellular survival and biofilm formation that predict eradication failure. However, this requires a larger and preferably prospective data set.
In conclusion, this explorative study showed a higher eradication failure rate in complicated MRSA carriers with ciprofloxacin-resistant MRSA lineages, which are predominantly healthcare-associated. In contrast, carriers of livestock-associated MRSA and the major community-associated ST8 and ST6 lineages were generally successfully decolonized. Further studies are warranted to confirm the higher eradication failure risk of ciprofloxacin-resistant lineages, and identify the underlying mechanisms. The identification of lineages that are prone to eradication failure is of clinical relevance, since it could influence the initiation and monitoring of MRSA eradication therapy.
Data Availability
The MRSA sequence data have been deposited in ENA under project number PRJEB59407.
References
Antimicrobial Resistance Collaborators (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399(10325):629–655
Davis KA et al (2004) Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis 39(6):776–782
Turner NA et al (2019) Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17(4):203–218
WHO Regional Office for Europe, European Centre for Disease Prevention and Control (2022) Antimicrobial resistance surveillance in Europe 2022–2020 data. WHO Regional Office for Europe, Copenhagen
Ammerlaan HS et al (2011) Eradication of carriage with methicillin-resistant Staphylococcus aureus: effectiveness of a national guideline. J Antimicrob Chemother 66(10):2409–2417
Huang SS et al (2019) Decolonization to reduce postdischarge infection risk among MRSA carriers. N Engl J Med 380(7):638–650
Westgeest AC et al (2021) Complicated carriage with methicillin-resistant Staphylococcus aureus: evaluation of the effectiveness of decolonization regimens advised in the Dutch National Guideline. Antimicrob Agents Chemother 65(9):e0025721
Holm MKA et al (2022) Estimated roles of the carrier and the bacterial strain when methicillin-resistant Staphylococcus aureus decolonization fails: a case-control study. Microbiol Spectr 10:e0129622
Kang HM et al (2021) Mupirocin and chlorhexidine genotypic resistance found in Staphylococcus aureus Isolated from young infants below 90 days old: a genetic basis for eradication failure. Pediatr Infect Dis J 40(1):49–54
Otto M (2012) MRSA virulence and spread. Cell Microbiol 14(10):1513–1521
Cheung GYC, Bae JS, Otto M (2021) Pathogenicity and virulence of Staphylococcus aureus. Virulence 12(1):547–569
Hetem DJ et al (2013) Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus. Emerg Infect Dis 19(11):1797–1802
Humphreys H, Coleman DC (2019) Contribution of whole-genome sequencing to understanding of the epidemiology and control of meticillin-resistant Staphylococcus aureus. J Hosp Infect 102(2):189–199
Laabei M et al (2014) Predicting the virulence of MRSA from its genome sequence. Genome Res 24(5):839–849
Bagge K et al (2019) Eradicating MRSA carriage: the impact of throat carriage and Panton-Valentine leukocidin genes on success rates. Eur J Clin Microbiol Infect Dis 38(4):683–688
Dutch Working Party on Antibiotic Policy (Stichting Werkgroep Antibiotica Beleid [SWAB]) (2012) Guideline for the treatment of MRSA carriage. Secretariaat SWAB, Nijmegen, the Netherlands
Bernards AT, Bonten MJM, Cohen Stuart J, Diederen BMW, Goessens WHF, Grundmann H, Kluytmans JAJW, Kluytmans-van den Bergh MFQ, Leversteinvan Hall MA, Mouton JW, Al Naiemi N, Troelstra A, Vandenbroucke-Grauls CMJE, Vos MC, Voss A (2012) NVMM Guideline Laboratory detection of highly resistant microorganisms (HRMO), version 2.0. Netherlands Society for Medical Microbiology, Leeuwarden, the Netherlands. https://www.nvmm.nl/media/1051/2012_hrmo_mrsa_esbl.pdf
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. http://www.eucast.org
Leopold SR et al (2014) Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52(7):2365–2370
Strauß L et al (2016) Detecting Staphylococcus aureus virulence and resistance genes: a comparison of whole-genome sequencing and DNA microarray technology. J Clin Microbiol 54(4):1008–1016
Wettstein Rosenkranz K et al (2014) Nasal carriage of methicillin-resistant Staphylococcus aureus (MRSA) among Swiss veterinary health care providers: detection of livestock- and healthcare-associated clones. Schweiz Arch Tierheilkd 156(7):317–325
Lindsay JA (2013) Hospital-associated MRSA and antibiotic resistance-what have we learned from genomics? Int J Med Microbiol 303(6–7):318–323
Fuzi M, Szabo D, Csercsik R (2017) Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front Microbiol 8:2261
Ortiz-Padilla M et al (2020) Role of low-level quinolone resistance in generating tolerance in Escherichia coli under therapeutic concentrations of ciprofloxacin. J Antimicrob Chemother 75(8):2124–2132
Brauner A et al (2016) Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14(5):320–330
Lechner S et al (2012) Interplay between population dynamics and drug tolerance of Staphylococcus aureus persister cells. J Mol Microbiol Biotechnol 22(6):381–391
Jörgensen J et al (2018) The majority of MRSA colonized children not given eradication treatment are still colonized one year later: systemic antibiotics improve the eradication rate. Infect Dis 50(9):687–696
Rieg S et al (2013) Microarray-based genotyping and clinical outcomes of Staphylococcus aureus bloodstream infection: an exploratory study. PLoS ONE 8(8):e71259
Acknowledgements
The authors would like to thank Kees Swenne and Karin Wold for their invaluable help with data collection.
Funding
This project was partially funded by the Antibiotic Resistance Network Holland West.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interests to declare.
Ethical Approval
Approval and patient consent were waived by the medical ethical committee of the University Medical Center Groningen.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Westgeest, A.C., Schippers, E.F., Rosema, S. et al. Genetic Determinants in MRSA Carriage and Their Association with Decolonization Outcome. Curr Microbiol 81, 63 (2024). https://doi.org/10.1007/s00284-023-03581-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03581-w