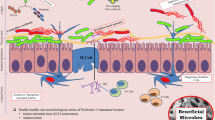
Abstract
In this study, newly isolated lactic acid bacteria (LAB) were screened for the potential bioremediation capacity against toxic lead (Pb, II) and cadmium (Cd, II) with their bioaccessibility and survivability. Five strains were selected from eighteen previously isolated probiotic LAB strains based on heavy metal-resistant potentiality through in vitro disc-diffusion assay. These five strains were evaluated in vitro to explore the Pb and Cd binding and removal efficiencies using Flame Atomic Absorption Spectrophotometry. At the same time, their bioaccessibility and survivability were assessed in a dynamic in vitro gastrointestinal digestion model. The results revealed that all the tested strains were shown to have a high magnitude of minimum inhibitory concentration values ranging from 500 to 2000 mg/L with 5 to 25 mm growth inhibition zones. The results also demonstrated a significant (P < 0.05) removal of Pb and Cd among five tested LAB strains. Lactobacillus delbrueckii subsp. bulgaricus LDMB02 showed the highest removal rates of Pb and Cd. It was also revealed that these strains significantly reduced Pb and Cd bioaccessibility from 42 to 50% and 40 to 58%, respectively. Moreover, these strains were shown to have significant survivability against Pb and Cd, ranging from 80.1 to 85.4% and 81.5 to 87.5%, respectively. This study recommends the immense potential exploit of LAB as a probiotic to protect human health from the adverse effects of Pb and Cd toxicity.




Similar content being viewed by others
Data Availability
Data and material of this research are accessible from the corresponding authors upon rational request.
Code Availability
Not applicable.
References
Pakdel M, Soleimanian-Zad S, Akbari-Alavijeh S (2019) Screening of lactic acid bacteria to detect potent biosorbents of lead and cadmium. Food Control 100:144–150. https://doi.org/10.1016/j.foodcont.2018.12.044
Dai QH, Bian XY, Li R, Jiang CB, Ge JM, Li BL, Ou J (2019) Biosorption of lead (II) from aqueous solution by lactic acid bacteria. Water Sci Technol 79:627–634. https://doi.org/10.2166/wst.2019.082
Elsanhoty RM, Al-Turki IA, Ramadan MF (2016) Application of lactic acid bacteria in removing heavy metals and aflatoxin B1 from contaminated water. Water Sci Technol 74:625–638. https://doi.org/10.2166/wst.2016.255
Bhakta JN, Munekage Y, Ohnishi K, Jana BB (2012) Isolation and identification of cadmium-and lead-resistant lactic acid bacteria for application as metal removing probiotic. Int J Environ Sci Technol 9:433–440. https://doi.org/10.1007/s13762-012-0049-3
ATSDR (2015) Agency for Toxic Substances and Disease Registry. CERCLA Priority List of Hazardous Substances, U.S. Department of Health and Human Services, Atlanta
Järup L (2003) Hazards of heavy metal contamination. Br Med Bull 68:167–182. https://doi.org/10.1093/bmb/ldg032
JrJ H (2016) Development of an inhalation unit risk factor for cadmium. Regul Toxicol Pharmacol 77:175–183. https://doi.org/10.1016/j.yrtph.2016.03.003
Nordberg GF, Fowler BA, Nordberg M (eds) (2014) Handbook on the toxicology of metals. Academic press, Cambridge
Akinci G, Guven DE (2011) Bioleaching of heavy metals contaminated sediment by pure and mixed cultures of Acidithiobacillus spp. Desalination 268:221–226. https://doi.org/10.1016/j.desal.2010.10.032
Rawat AP, Giri K, Rai JPN (2014) Biosorption kinetics of heavy metals by leaf biomass of Jatropha curcas in single and multi-metal system. Environ Monit Assess 186:1679–1687. https://doi.org/10.1007/s10661-013-3485-8
Balooch FD, Fatemi SJ, Iranmanesh M (2014) Combined chelation of lead (II) by deferasirox and deferiprone in rats as biological model. Biometals 27:89–95. https://doi.org/10.1007/s10534-013-9689-0
Porova N, Botvinnikova V, Krasulya O, Cherepanov P, Potoroko I (2014) Effect of ultrasonic treatment on heavy metal decontamination in milk. Ultrason Sonochem 21:2107–2111. https://doi.org/10.1016/j.ultsonch.2014.03.029
Mrvčić J, Stanzer D, Šolić E, Stehlik-Tomas V (2012) Interaction of lactic acid bacteria with metal ions: opportunities for improving food safety and quality. World J Microbiol Biotechnol 28:2771–2782. https://doi.org/10.1007/s11274-012-1094-2
Feng M, Chen X, Li C, Nurgul R, Dong M (2012) Isolation and identification of an exopolysaccharide-producing lactic acid bacterium strain from Chinese Paocai and biosorption of Pb (II) by its exopolysaccharide. J Food Sci 77:T111–T117. https://doi.org/10.1111/j.1750-3841.2012.02734.x
Li PS, Tao HC (2015) Cell surface engineering of microorganisms towards adsorption of heavy metals. Crit Rev Microbiol 41(140):149. https://doi.org/10.3109/1040841X.2013.813898
Jadán-Piedra C, Alcántara C, Monedero V, Zúñiga M, Vélez D, Devesa V (2017) The use of lactic acid bacteria to reduce mercury bioaccessibility. Food Chem 228:158–166. https://doi.org/10.1016/j.foodchem.2017.01.157
Zhai Q, Yin R, Yu L, Wang G, Tian F, Yu R, Chen W (2015) Screening of lactic acid bacteria with potential protective effects against cadmium toxicity. Food Control 54:23–30. https://doi.org/10.1016/j.foodcont.2015.01.037
Yi YJ, Lim JM, Gu S, Lee WK, Oh E, Lee SM, Oh BT (2017) Potential use of lactic acid bacteria Leuconostocmesenteroides as a probiotic for the removal of Pb (II) toxicity. J Microbiol 55:296–303. https://doi.org/10.1007/s12275-017-6642-x
Bhakta JN, Ohnishi K, Munekage Y, Iwasaki K, Wei MQ (2012) Characterization of lactic acid bacteria-based probiotics as potential heavy metal sorbents. J Appl Microbiol 112:1193–1206. https://doi.org/10.1111/j.1365-2672.2012.05284.x
Zhai Q, Xiao Y, Zhao J, Tian F, Zhang H, Narbad A, Chen W (2017) Identification of key proteins and pathways in cadmium tolerance of Lactobacillus plantarum strains by proteomic analysis. Sci Rep 7:1–17. https://doi.org/10.1038/s41598-017-01180-x
Ojekunle O, Banwo K, Sanni AI (2017) In vitro and in vivo evaluation of Weissellacibaria and Lactobacillus plantarum for their protective effect against cadmium and lead toxicities. Lett Appl Microbiol 64:379–385. https://doi.org/10.1111/lam.12731
Muhammad Z, Ramzan R, Zhang R, Zhao D, Gul M, Dong L, Zhang M (2021) Assessment of in vitro and in vivo bioremediation potentials of orally supplemented free and microencapsulated Lactobacillus acidophilus KLDS strains to mitigate the chronic lead toxicity. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2021.698349
Li B, Jin D, Yu S, Etareri-Evivie S, Muhammad Z, Huo G, Liu F (2017) In vitro and in vivo evaluation of Lactobacillus delbrueckii subsp. bulgaricus KLDS1.0207 for the alleviative effect on lead toxicity. Nutrients 9:845. https://doi.org/10.3390/nu9080845
Muhammad Z, Ramzan R, Zhang S, Hu H, Hameed A, Bakry AM, Pan S (2018) Comparative assessment of the bioremedial potentials of potato resistant starch-based microencapsulated and non-encapsulated Lactobacillus plantarum to alleviate the effects of chronic lead toxicity. Front Microbiol 9:1306. https://doi.org/10.3389/fmicb.2018.01306
Al-Wabel NA, Mousa HM, Omer OH, Abdel-Salam AM (2007) Biological evaluation of synbiotic fermented milk against lead acetate contamination in rats. J Food Agric Environ 5:169
Jafarpour D, Shekarforoush SS, Ghaisari HR, Nazifi S, Sajedianfard J, Eskandari MH (2017) Protective effects of synbiotic diets of Bacillus coagulans, Lactobacillus plantarum and inulin against acute cadmium toxicity in rats. BMC Complement Altern Med 17:1–8. https://doi.org/10.1186/s12906-017-1803-3
Topcu A, Bulat TUĞBA (2010) Removal of cadmium and lead from aqueous solution by Enterococcus faecium strains. J Food Sci 75:T13–T17. https://doi.org/10.1111/j.1750-3841.2009.01429.x
Halttunen T, Collado MC, El-Nezami H, Meriluoto J, Salminen S (2008) Combining strains of lactic acid bacteria may reduce their toxin and heavy metal removal efficiency from aqueous solution. Lett Appl Microbiol 46:160–165. https://doi.org/10.1111/j.1472-765X.2007.02276.x
Li X, Ming Q, Cai R, Yue T, Yuan Y, Gao Z, Wang Z (2020) Biosorption of Cd2+ and Pb2+ from apple juice by the magnetic nanoparticles functionalized lactic acid bacteria cells. Food Control 109:106916. https://doi.org/10.1016/j.foodcont.2019.106916
Massoud R, Khosravi-Darani K, Sharifan A, Asadi G, Zoghi A (2020) Lead and cadmium biosorption from milk by Lactobacillus acidophilus ATCC 4356. Food Sci Nutr 8:5284–5291. https://doi.org/10.1002/fsn3.1825
Islam MZ, Uddin ME, Rahman MT, Islam MA, Harun-ur-Rashid M (2021) Isolation and characterization of dominant lactic acid bacteria from raw goat milk: assessment of probiotic potential and technological properties. Small Rumin Res 205:106532. https://doi.org/10.1016/j.smallrumres.2021.106532
Kang CH, Kwon YJ, So JS (2016) Bioremediation of heavy metals by using bacterial mixtures. Ecol Eng 89:64–69. https://doi.org/10.1016/j.ecoleng.2016.01.023
Kumar N, Kumar V, Panwar R, Ram C (2017) Efficacy of indigenous probiotic Lactobacillus strains to reduce cadmium bioaccessibility-an in vitro digestion model. Environ Sci Pollut Res 24:1241–1250. https://doi.org/10.1007/s11356-016-7779-6
Islam MZ, Tabassum S, Harun-ur-Rashid M, Vegarud GE, Alam MS, Islam MA (2021) Development of probiotic beverage using whey and pineapple (Ananas comosus) juice: sensory and physico-chemical properties and probiotic survivability during in-vitro gastrointestinal digestion. J Agric Food Res 4:100144. https://doi.org/10.1016/j.jafr.2021.100144
Li K, Ramakrishna W (2011) Effect of multiple metal resistant bacteria from contaminated lake sediments on metal accumulation and plant growth. J Hazard Mater 189:531–539. https://doi.org/10.1016/j.jhazmat.2011.02.075
Shu G, Zheng Q, Chen L, Jiang F, Dai C, Hui Y, Du G (2021) Screening and identification of Lactobacillus with potential cadmium removal and its application in fruit and vegetable juices. Food Control 126:108053. https://doi.org/10.1016/j.foodcont.2021.108053
Qing HU, Dou MN, Qi HY, Xie XM, Zhuang GQ, Min YANG (2007) Detection, isolation, and identification of cadmium-resistant bacteria based on PCR-DGGE. J Environ Sci 19:1114–1119. https://doi.org/10.1016/S1001-0742(07)60181-8
Chang YC, Choi D, Kikuchi S (2012) Enhanced extraction of heavy metals in the two-step process with the mixed culture of Lactobacillus bulgaricus and Streptococcus thermophilus. Bioresour Technol 103:477–480. https://doi.org/10.1016/j.biortech.2011.09.059
George F, Mahieux S, Daniel C, Titécat M, Beauval N, Houcke I, Garat A (2021) Assessment of Pb (II), Cd (II), and Al (III) removal capacity of bacteria from food and gut ecological niches: insights into biodiversity to limit intestinal biodisponibility of toxic metals. Microorganisms 9:456. https://doi.org/10.3390/microorganisms9020456
Halttunen T, Salminen S, Tahvonen R (2007) Rapid removal of lead and cadmium from water by specific lactic acid bacteria. Int J Food Microbiol 114:30–35. https://doi.org/10.1016/j.ijfoodmicro.2006.10.040
Le B, Yang SH (2019) Biosorption of cadmium by potential probiotic Pediococcuspentosaceus using in vitro digestion model. Biotechnol Appl Biochem 66:673–680. https://doi.org/10.1002/bab.1783
Sun L, Liu G, Yang M, Zhuang Y (2012) Bioaccessibility of cadmium in fresh and cooked AgaricusblazeiMurill assessed by in vitro biomimetic digestion system. Food Chem Toxicol 50:1729–1733. https://doi.org/10.1016/j.fct.2012.02.044
Yang LS, Zhang XW, Li YH, Li HR, Wang Y, Wang WY (2012) Bioaccessibility and risk assessment of cadmium from uncooked rice using an in vitro digestion model. Biol Trace Elem Res 145:81–86. https://doi.org/10.1007/s12011-011-9159-x
Amiard JC, Amiard-Triquet C, Charbonnier L, Mesnil A, Rainbow PS, Wang WX (2008) Bioaccessibility of essential and non-essential metals in commercial shellfish from Western Europe and Asia. Food Chem Toxicol 46:2010–2022. https://doi.org/10.1016/j.fct.2008.01.041
Kabak B, Ozbey F (2012) Aflatoxin M1 in UHT milk consumed in Turkey and first assessment of its bioaccessibility using an in vitro digestion model. Food Control 28:338–344. https://doi.org/10.1016/j.foodcont.2012.05.029
Serrano-Niño JC, Cavazos-Garduño A, Hernandez-Mendoza A, Applegate B, Ferruzzi MG, San Martin-González MF, García HS (2013) Assessment of probiotic strains ability to reduce the bioaccessibility of aflatoxin M1 in artificially contaminated milk using an in vitro digestive model. Food Control 31:202–207. https://doi.org/10.1016/j.foodcont.2012.09.023
Daisley BA, Monachese M, Trinder M, Bisanz JE, Chmiel JA, Burton JP, Reid G (2019) Immobilization of cadmium and lead by Lactobacillus rhamnosus GR-1 mitigates apical-to-basolateral heavy metal translocation in a Caco-2 model of the intestinal epithelium. Gut Microbes 10:321–333. https://doi.org/10.1080/19490976.2018.1526581
Argyri AA, Zoumpopoulou G, Karatzas KAG, Tsakalidou E, Nychas GJE, Panagou EZ, Tassou CC (2013) Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol 33:282–291. https://doi.org/10.1016/j.fm.2012.10.005
Mulaw G, SisayTessema T, Muleta D, Tesfaye A (2019) In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int J Microbiol 2019:1–11. https://doi.org/10.1155/2019/7179514
Banwo K, Sanni A, Tan H (2013) Technological properties and probiotic potential of Enterococcus faecium strains isolated from cow milk. J Appl Microbiol 114:229–241. https://doi.org/10.1111/jam.12031
Acknowledgements
The authors acknowledge the Ministry of Science and Technology for their financial support. The authors also thank the Dairy Microbiology and Biotechnology Lab, Department of Dairy Science, and Food Science Lab, Interdisciplinary Institute for Food Security, Bangladesh Agricultural University, Mymensingh, for their necessary facilities.
Funding
This work was supported by National Science and Technology (NST) fellowship (298, 2019-2020), Ministry of Science and Technology, Dhaka, Bangladesh.
Author information
Authors and Affiliations
Contributions
MSH: Formal investigation, analysis, writing first & original draft, review & edited final manuscript. MZI: Conceptualization & study design, set-up & optimized biomimetic digestion model, formal investigation, data curation & analysis, graphical presentation, writing original draft and review & edited final manuscript. RIL: Review and manuscript proofreading. MAHS: Review & edited the draft & final manuscript. MAI: Supervision, technical support, review & edited final manuscript. MHR: Supervision and allocation of lab resources for this research work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared no potential conflicts of interest concerning this article's research, authorship, and/or publication.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2022_3059_MOESM1_ESM.tiff
Supplementary file1 (TIFF 14301 kb). Figure S1. Growth inhibition zones at different Pb and Cd concentrations on MRS agar media (a) Lactobacillus acidophilus LDMB01; (b) Lactobacillus delbrueckii subsp. bulgaricus LDMB02; (c) Lacticaseibacillus casei subsp. casei LDMB03; (d) Pediococcus pentosaceus LDMB08; and (e) Lactococcus lactis LDMB10. A, B and C indicate the heavy metal concentrations at 50, 250, and 2000 mg/L, respectively. The plates are labeled with Pb = Lead and Cd = Cadmium, and the disc without labeling indicates the control group
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hasan, M.S., Islam, M.Z., Liza, R.I. et al. Novel Probiotic Lactic Acid Bacteria with In Vitro Bioremediation Potential of Toxic Lead and Cadmium. Curr Microbiol 79, 387 (2022). https://doi.org/10.1007/s00284-022-03059-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03059-1