Abstract
Polyhydroxyalkanoates (PHA) being biological polymers have attracted great attention. PHA have similar properties to that of synthetic plastic and are biodegradable. To discourage plastic pollution in the environment alternative solutions to the plastic pollution has to be readily available. High cost in production of PHA limits the production of these polymers at industrial scale. Bacteria are screened for PHA from diverse niches to meet the current requirements of cheap PHA production at industrial level. The microbial biofilm formed on the surface of microplastic could be a potential source in providing bacteria of economic importance. This paper is an attempt to search microplastic niche for potential PHA producers. PHA production variation was observed with different parameters such as type of carbon source, nitrogen source concentration and also time of incubation. Bacillus sp. CM27 showed maximum PHA yield up to 32.1% among other isolates at 48 h with 2% glucose as carbon source. Optimization of media leads to increase in PHA yield (37.69%) of CDW in Bacillus sp. CM27. Amino acid sequence of Bacillus sp.CM27 showed the presence of PhaC box with sequence, G-Y-C-M-G-G having cysteine in the middle of the box. The extracted polymer was confirmed by FTIR spectroscopy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plastic production accelerated after its commercial development in 1930 and 1940s as it turned out to be a convenient and cheap material in use. Packaging materials developed for immediate disposal have taken a major part of market sector for plastic resins. Due to recent COVID-19 pandemic, plastic waste generation has spiked adding to existing plastic waste pollution [1]. Plastic pollution is a global threat damaging aquatic as well as terrestrial life forms which have been listed among existing threats such as climate change, ocean acidification and ozone depletion [2]. Open disposal of plastic without systematic waste management has constrained UN Sustainable Development Goals achievements [3]. To combat rising pollution due to plastic use, alternative material such as bioplastics obtained from PHA, accounting for complete degradation into carbon dioxide and water [4], is required. Polyhydroxybutyrate (PHB) is a common type of polyhydroxyalkanoates having similar properties to that of conventional plastics such as polypropylene and polyethylene. Polyhydroxyalkanoates are synthesized by bacteria as organic inclusion granules that represent as complex class of storage polyesters mainly composed of hydroxy fatty acids in the presence of high concentration of carbon source and limiting one of the essential nutrients such as nitrogen, phosphorus, oxygen or sulphur. The discovery of PHB in Bacillus megaterium bacterium leads the scope in finding different types of polyhydroxyalkanoate in bacterial species. Over 90 bacterial genera are being marked to accumulate PHB and related PHA intracellularly in gram-negative and gram-positive bacteria [5]. Bacteria from genus Bacillus are gaining attention in terms of industrial use for PHA production due to its capability of producing endotoxin-free PHA, beneficial for applications in medical field.
PHA commercialization is deviating from the expected market trend due to its high cost of production on an industrial scale, which raises the cost of bioplastic in comparison with conventional plastic. Isolation of suitable bacterial strain is the key starting point in PHA production at industrial scale as the polymer produced can differ from 30 to 80% of the cell dry weight (CDW) based on the strain and culture conditions [6]. Marine bioprospecting may lead to exploration of potential PHA producers as marine bacteria are not much explored [7].
In the ocean, plastic debris is considered a novel type of floating substratum for microbial attachment and transportation over a long distance as this plastic lasts longer than any other natural substrates like macroalgae, feathers or wood. [8]. Microplastics are fragments of plastic marine debris that are less than 5 mm [9]. The surface of plastic promotes microbial biofilms due to its hydrophobic nature [8]. Increased microplastic abundance in the ocean due to an extrapolated increase in plastic pollution has aided microbial biofilm formation on this artificial substrate. Microplastics provide carbon sources to microbes that can catalyse metabolic reactions which contribute to microplastic adsorption, desorption and breakdown [10, 11]. Therefore, a new microbial niche is furnished by microplastics in the marine environment, having microbial diversity associated with the surface of microplastics more or less differing from free-living or other particles-associated in the surrounding water. [12]. The goal of this research was to look for bacteria potentially producing PHA intracellularly from the surface of microplastics collected from marine debris at seashore. PHA production within bacteria is influenced by external environmental conditions such as carbon source type, time required for PHA production, and high carbon and nitrogen-limited conditions. By maintaining the culture conditions, the objective was to get the maximum PHA-producing bacteria among the isolates. Currently, there are few reports on bacteria isolated from biofilms obtained from the surface of microplastics [13, 14].
Materials and Methods
Sample Collection
Microplastic pellets were searched and retrieved from the debris collected on shore due to tides action with sterile forceps. The Caranzalem beach was selected as location site (15° 27′ 59.4ʺ N, 73° 48′ 17.1 ʺ E) and microplastic pellets were transported under sterile condition.
Isolation, Identification and Screening of PHA-Producing Bacteria
Microplastic pellets were rinsed with sterile distilled water to remove the topsoil layer. The microplastic pellets were vortexed with sterile saline and serial dilutions were carried out. 100 μL of inoculum is plated on Zobell Marine Agar (ZMA) medium (Himedia) and incubated for 24 h at 30 °C. Based on morphological differences, each colony was picked and purified by quadrant streaking.
Marine basal medium (sea salt = 2.8 g/100 mL, tryptone broth = 0.25 g/100 mL, yeast extract = 0.1 g/100 mL) [15], and modified E2 media (NaNH4HPO4.4H2O = 0.35 g/100 mL, K2HPO4 = 0.5 g/100 mL, KH2PO4 = 0.35 g/100 mL, MgSO4 = 100 mM, 100 μL of MT microelement stock solution) plates were used for screening of PHA-producing bacteria by using 2% glucose as carbon source. Isolated bacterial colonies were grown on these plates for 48-h’ incubation at 30 °C. After incubation, the plates were screened for intracellular lipid by using 0.02% of ethanolic solution of Sudan Black B [16].
Bacteria stained positively with Sudan Black B were identified by molecular method as a potential PHA producer. Bacterial DNA was isolated by Genomic Extraction Kit (Sigma Aldrich) method. Bacterial DNA was amplified by using 16 S rRNA universal primers (forward 5′-GAGAGTTTGATCCTGGCTCAG-3′ (27 F) and reverse primer 5′-TACGGYTACCTTGTTACGACTT-3′ (1492R) by polymerase chain reaction. Total 50 μL of reaction volume was ran as PCR mixture containing 3 μL DNA template, 3 μL of forward and reverse primers each, 25 μL of Master mix (promega), and 16 μL of distilled water. The thermal cycling program was carried out with an initial denaturation at 94 °C for 4 min continued by keeping final denaturation at 94 °C for 1 min with an annealing temperature at 55 °C for 1 min, 72 °C for 1 min which was of 35 cycles and final extension at 72 °C for 20 min. The amplified PCR products were confirmed by gel electrophoresis method. Sanger’s dideoxynucleotide chain termination method was used for sequencing after purification of PCR product. The sequencing was carried out at Bioserve Biotechnologies India Pvt Ltd. The similarity of the 16S rRNA gene sequences obtained was checked with the bacterial sequences available at National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast) using Basic Local Alignment Search Tool (BLAST) program. Mega 6 software was used to carry out phylogenetic and evolutionary analysis by means of maximum likelihood method.
PHA Production
The selected bacterial strains were grown first in Zobell marine broth (Himedia) and incubated at 30 °C for 24 h. OD was taken at 600 nm. 2 mL of culture was inoculated in 100 mL of marine basal medium media (MBM) with 2% of glucose as carbon source. The culture was collected in centrifuge tube at 48 and 72 h, respectively, and 5 ml was centrifuged at 8000 rpm for 10 min with subsequent sterile distilled water to remove traces of broth. The cell pellet was dried and treated with 5 ml of 4% sodium hypochlorite followed by incubation at 37 °C for 20 min. Centrifugation was carried out at 8000 rpm for 10 min. Later pellet was washed with cold diethyl ether [17]. The pellet is dissolved in hot chloroform and 0.5 mL overnight dried sample is further assayed by Slepecky and Law’s method [18] for PHB quantification. The standard graph was prepared of PHB obtained from sigma in concentration of 1 mg/mL. The selected bacterial strains for PHA production were grown and quantified in triplicates.
Optimization of Culture Growth and Media for PHA Production
Time Course of Growth
The time taken for growth and the production of PHA by bacterial culture CM27 was studied. 1% of pre-grown culture was used to inoculate 100 ml of MBM medium at 30 °C for 150 rpm. After every 24 h’ interval, 5 mL broth was removed during cultivation period up to day 4 and centrifugation was carried out at 8000 rpm for 10 min. The dried pellet was then assayed by Slepecky and Law’s method [18]. The experiment was conducted in triplicates.
Optimization of Culture Media for PHA Production
Optimization was carried out by changing one factor at a time. The carbon source was changed in MBM by keeping constant nitrogen source. In 250-mL flask, 100 ml broth was prepared with 2% glucose, xylose and sucrose, respectively. 2 mL of pre-grown culture was inoculated in each flask containing this media. After 48 h of incubation at 30 °C with 150 rpm, 5 mL of culture was centrifuged and further dried pellet was assayed by Slepecky and Law’s method. After changing carbon source, nitrogen source was changed sequentially. In 100 mL MBM broth, yeast extract concentration in the range 0.5–1.5 g/L was taken in each flask with 2% glucose and 2.5 g/L tryptone broth. 2 mL of pre-grown culture was inoculated in each flask containing this media. After 48 h of incubation at 30 °C with 150 rpm. 5 mL of culture was centrifuged and further dried pellet was assayed by Slepecky and Law’s method. Later tryptone broth concentration was changed in the range of 1.5 to 3.5 g/L with 0.5 g/L difference. In 100 mL MBM broth, 2% glucose and 1 g/L yeast extract were taken as optimized parameters with changing tryptone broth concentration. Similarly, 2 mL of pre-grown culture was inoculated and after 48 h of incubation at 30 °C with 150 rpm, PHA quantification was carried out as described earlier. Optimization experiments were carried out in triplicates.
Primer Selection, Amplification and Sequencing of PHA Synthase Gene
According to the published sequences of PHA synthase, gene specific primers are reported by T.R. Shamala and coworkers [19] for Bacillus spp. The primers B1F (AACTCCTGGGCTTGAAGACA) and B1R (TCGGAATATGATCACGGCTA) were used to amplify PHA synthase gene from bacterial strain CM27 by polymerase chain reaction. The reaction mixture consisted of template (3 μL), forward primer (3 μL), reverse primer (3 μL), taq polymerase mixture (25 μL), and water (16 μL), the total volume was 50 μL. The thermal cycler program was carried on applied biosystem thermal cycler with following conditions, Initial denaturation at 95 °C for 5 min and 40 cycles of 95 °C for 1 min, 52 °C for 45 s 72 °C for 1 min with final extension at 72 °C for 10 min by following incubation at 4 °C. The amplified PCR products were confirmed by gel electrophoresis by visualizing DNA bands under UV transilluminator and the analysed PCR products by Bioserve Biotechnologies India Pvt Ltd were converted to fasta format. The sequences were blasted on NCBI blast for similarity by blastx. The sequences were submitted to NCBI for Accession number.
FTIR Analysis
The PHA extracted from CM27 culture was analysed by FTIR spectroscopy. Shimadzu FTIR Affinity-1 spectrophotometer was used with spectral range, 4000–400 cm−1, to record the IR spectra.
Statistical Analysis
Each sample was run in triplicates and standard deviation was determined. The data obtained were analysed by one-way ANOVA using Microsoft excel software to determine the significance.
Results
Isolation and Screening of PHA-Producing Bacteria
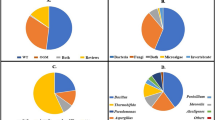
Microplastic pellets were collected from the shoreline on Caranzalem beach, Goa, India. In total, 30 bacterial strains were isolated from the surface of microplastic pellets based on morphological characteristics using Zobell Marine agar. These isolates were screened for intracellular lipids by staining with Sudan Black B; of these, 6 isolates were positively stained. The negatively stained isolates were discarded considering their inability to produce PHA. The positively stained isolates were then cultured and identified using16S rRNA gene amplification product which was around 1500 bp. Nucleotide sequences, procured from Sanger’s dideoxy method (Analysis were carried by Bioserve), were blasted on NCBI Nucleotide Blast tool for bacterial identification, considering the hits showing maximum similarity. The nucleotide sequences were deposited to GenBank database with Accession numbers MT879600.2, MT879601.2, MT879602.2, MT879603.2, MW397191.2 and MT907456.2. Two bacterial strains were of genus Bacillus, two bacterial strains were of genus Priestia and two bacterial strains were of genus Pseudomonas. The phylogenetic tree was constructed as shown in Fig. 1. by Mega 7 software with maximum likelihood method.
The phylogenetic tree of PHA-producing bacteria isolated from microplastic and its nearest neighbours derived from the maximum likelihood method based on the partial sequence of the 16S rRNA gene. GenBank accession numbers are indicated in parenthesis. Type strains are used as reference strains with superscript T. Bootstrap values (based on 1000 replication) > 50% are shown at branch points. Bar, 0.05 means the nucleotide substitution per position
Selection of Bacterial Isolate with High PHA Accumulation
Quantitative estimation of PHA granules was carried out by measuring PHB at 235 nm after converting it to crotonic acid with concentrated sulphuric acid by maintaining heating condition at 100 °C for 10 min. Bacillus sp. CM27 (MW397191.2) showed highest PHB accumulation than the other isolated strains in Marine Basal medium (MBM) media with PHA yield maximum (32.2% per CDW) at 48 h of incubation than 72 h of incubation. PHB accumulation in CM36 was 26.29% for 48 h and decreased to 11.42% for 72 h. The least PHB accumulation was seen in CM23 and CM26 that is in the range of 0.9–2.5% for 48 and 72 h. PHB accumulation in CM3 and CM10 was 25.08 and 22.22%, respectively, for 48 h which decreased to 23.63 and 20.61% for 72 h, respectively. Among the six strains of bacteria, CM27 was selected for further optimization which is depicted in Fig. 2. By using one-way ANOVA, there is significant difference in PHA production by isolated bacteria wherein p value is less than F critical value thus rejecting null hypothesis.
The selected screened isolates accumulating PHA grown in MBM media with 2% glucose for 48 and 72 h of incubation. PHA content in bacteria was tested by Slepecky and Law’s method wherein crotonic acid is measured at 235 nm. The results are averages of ± standard deviations from three separate experiments. *Statistical analysis was carried out by one-way ANOVA test, *p value (≤ 0.05)
Optimizing Nutrient Composition of Marine Basal Medium and Incubation Period for Increased PHA Production
Incubation Period
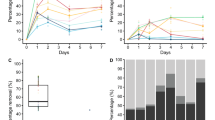
In this study, the experiments were carried out by keeping sea salt and carbon source (2%) concentration constant in the given medium. The strain CM27 identified as Bacillus sp. was grown in MBM broth and was sampled at regular time interval that is from 24 to 96 h. It was observed that PHA accumulation was high at 48 h and then decreased subsequently at 72 h and 96 h from 32.1% to 28.44% and 26.55%, respectively, as shown in Fig. 3.a. Statistical method denoted that there is significant difference in PHA production with time with p value < 0.05.

source on PHA production in Bacillus sp. CM27. Fig. c.,Effect of Yeast Extract Concentration on PHA production in Bacillus sp. CM27. Fig. d, Effect of Tryptone Concentration on PHA production Bacillus sp. CM27. Analysis was carried out by converting PHA to crotonic acid by Slepecky and Law’s method measured at 235 nm. The results are averages of ± standard deviations from three separate experiments. Fig. a, b and d *Statistical analysis was carried out by one-way ANOVA test, * p value (≤ 0.05). Fig. c. *Statistical analysis was carried out by one-way ANOVA test, *p value (= 0.08)
Effect on PHA production in Bacillus sp. CM27 during Optimizing incubation time and nutrient composition of Marine Basal Medium. Fig. a., Effect of Incubation condition on PHA production in Bacillus sp. CM27. Fig. b. Effect of different Carbon
Carbon Source
Among the carbon sources used for PHA production, glucose gave maximum PHA accumulation than sucrose and xylose. PHA yield with glucose was 32.79%, sucrose as 29.99% and xylose gave only 12.73% PHA accumulation in Bacillus sp. CM27 at 48 h of incubation time (Fig. 3.b.). There is a significant difference in PHA accumulation with different carbon source derived by one-factor ANOVA with p value < 0.05.
Nitrogen Source
The concentration of nitrogen source in the media was changed and PHA content was observed by keeping other components constant in the given media. As the yeast extract concentration increased, the PHA accumulation also increased up to certain concentration of yeast extract and with further increase in yeast extract in concentration PHA accumulation decreased. With 0.5 g/L concentration of yeast extract, PHA accumulation per CDW was 29.17% and it increased to 32.44% with 0.75 g/L. There was not much difference in PHA accumulation upon increasing yeast extract to 1 g/L which gave PHA yield as 32.8%. PHA accumulation decreased after the addition of 1.25 and 1.5 g/L of yeast extract to 26.42% and 26.61%, respectively (Fig. 3.c). The change in yeast extract concentration did not affect PHA production much as there was no significant difference with p value > 0.05 (p value = 0.08).
By keeping the optimum yeast extract concentration (1 g/L), further optimization of tryptone broth was carried out. It was observed that at 2 g/L of tryptone concentration, the Bacillus sp. CM27T gave maximum PHA accumulation which is about 37.69% of CDW. PHA accumulation with 1.5 g/L of tryptone broth was 6.41% which was least among other concentrations (Fig. 3.d). The subsequent increase in the amount of tryptone broth source from 2.5 g/L, 3 g/L and 3.5 g/L increased CDW with decrease in PHA yield. Tryptone broth concentration at 3.5 g/L in the medium decreased the PHA yield drastically to 23.96%. Thus, proving that adequate amount of nitrogen source proves to be beneficial for synthesis of PHA within Bacteria. The change in tryptone broth concentration showed a significant change in PHA production with p value < 0.05.
DNA Sequence Analysis of PHA Synthase Gene
The nucleotide sequence of PHA synthase gene was detected with an open reading frame of 354 bp encoding a protein with 118 amino acid residues. The pI and molecular mass of the protein were 4.7 and 13,285 Da, respectively. The PHA gene sequence of Bacillus sp. CM27 (GenBank No. MW810070) is 100% identical to the Bacillus sp. C19 PhaC gene sequence (ACJ63445.1.) as per the DDBJ/EMBL/GenBank databases. The catalytic domain of PHA synthase gene from Bacillus sp. CM27 showed extensive homology with PhaC subunits of the bacterial class III PHA synthases.
Amino Acid Composition of PHA Synthase Gene
Supplementary table 1 depicts the amino acid composition of the PHA synthase gene. In Bacillus sp. CM27 PHA synthase gene, relatively high abundance of Aspartic acid (D) 11.0%, Leucine (L) 10.2% was found. Analysis of the amino acid sequence of Bacillus sp. CM27 PHA synthase gene with other PHA synthase gene from bacterial isolates. The presence of the PhaC box sequence at the active site of catalytic subunit PhaC of PHA synthase gene is typically described as G-X-C-X-G-G (where X is an arbitrary amino acid), with cysteine as an active centre, PhaC box was observed in amino acid sequence of PHA synthase with PHA box sequence as G-Y-C-M-G-G (Fig: 4).
FTIR Analysis
The infrared spectra of polymer extracted from Bacillus sp. CM27 when grown in MBM media gave peak at 1733.12 cm−1 (Supplementary material Fig. 5b) representing bands of carbonyl (C=O) stretch confirming that the extracted polymer was PHA by the presence of functional group.
Discussion
The surface of plastic promotes microbial biofilms due to its hydrophobic nature [8] and with the increase in amount of microplastic in ocean due to plastic pollution, microorganisms are promoted to form biofilm on this artificial substrate. Thus, a new microbial niche is furnished by microplastic in the aquatic environment, and the microbial composition here vastly differs when compared with substrate-associated or free-living microbial diversity in the given environment [12]. We attempted to isolate bacteria from this novel substrate as it may provide a unique environment and we may get novel PHA-producing bacteria.
Microplastic collected from seashore harboured Bacillus and Pseudomanas bacterial genus with ability to produce PHA. Bacillus were dominant (66%) PHA producers. Bacillus species are always termed beneficial over other bacterial species in terms of PHA production as they lack lipopolysaccharide layer that makes extraction much easier [20]. Pseudomonas species in the presence of fatty acids produce mcl-PHAs whereas majority of these species examine so far are devoid of PHB synthesis ability [21].
In this study, Bacillus sp. CM27 showed high PHA accumulation (0.53 g/L) at 2% glucose concentration compared to sucrose (2%) and xylose (2%) in 48 h. Similar trend of PHA accumulation was observed in studies reported by Seid Mohammed and coworkers [21], where glucose produced higher amount of PHA than sucrose by using strains of Bacillus sp. BPPI-19 and Bacillus sp. BPPI-14. The concentration of glucose and sucrose reported here was 1% in Minimal Davis Broth (pH 7) and bacteria were grown for 72 h at 30 °C. PHA accumulation with glucose as a carbon source was 0.5 and 0.47 g/L and for sucrose, it was 0.2 and 0.19 g/L for Bacillus sp. BPPI-19 and Bacillus sp. BPPI-14, respectively. M.S. Sreekanth and coworkers [22] reported that glucose at concentration of 20 g/L yielded PHA around 0.444 g/L and sucrose yielded PHA production of 0.48 g/L by Bacillus sp. CFR 67. In the reports given by B.B. Salgaonkar [23], H16 strain of Bacillus megaterium isolated from solar salterns produced around 40% PHA per CDW at 42 h in E2 mineral media with 2% glucose. The reported study [24] says that during sucrose fermentation, Bacillus sp. JMa5 strain yielded 25–35%, (w/w) PHB. Bacillus subtilis 25 and Bacillus megaterium 21 with 2% glucose in nutrient broth yielded PHA around 19.51 and 19.49%, respectively, which was reported by Yüksekdağ, Z. N. [25].
Bacteria grown in excess carbon and reduced nitrogen source synthesize intracellular PHA in presence of high acetyl CoA, low free coenzyme A concentration and high NADPH. When bacteria are grown in nutrient rich medium, concentration of free coenzyme A is high and PHA synthesis is low [26].
It has been demonstrated that nutrient limitation triggers the production of mcl-PHAs [27]. Nitrogen or phosphorus limitation stimulated the synthesis of PHA in P. oleovorans growing on n-alkanes, P. putida BM01 and P. resinovorans on octanoate [28] and P. putida KT2442 on oleic acid [29]. Contradictory observation was noted where nutrient limitation was not useful for PHA synthesis in P. putida [30] and also in P. oleovorans [31] on octanoate. It appears that the relationship between growth conditions and PHA accumulation is determined by the single factor such carbon source, strain and cultivation conditions or a mixture of these factors [32]. High cell densities of Pseudomonas putida KT2440 was attained by using glucose as carbon source through automated feeding strategy [33].
In 2012, Sreekanth and coworkers reported that the quantity of PHA production increased by the incorporation of organic nitrogen source in production media than with inorganic nitrogen source. In their study, the concentration of nitrogen source as 0.2 g/L of tryptone yielded PHA as 0.012 g/L. Similarly, with yeast extract, PHA yield was 0.394 g/L. Protease peptone increased PHA yield of Bacillus subtilis 25 and Bacillus megaterium 21% to 78.69% and 77.00% when used as nitrogen source [26].
Kranz and coworkers [34] demonstrated that phaC gene is most important for synthesis of PHB by carrying out experiments with single and double mutation in phaA and phaAB that did not affect the PHA synthesis whereas single mutation in phaC prevented PHB production. Molecular methods for the detection of phaC gene that is assigned for encoding PHB synthase in the bacterial genome can be considered as confirmation of PHB production in the PHB-producing bacteria. In current study, the deduced amino acid sequence of the Bacillus sp.CM27T was matched with the other PHA synthase sequences in the NCBI database by means of BLAST search program, and it displayed high homology with PHA synthase class III, PhaC subunit from Bacillus sp. C19 (GenBank Accession No. ACJ63445.1).
The PhaC subunit is reported to have a lipase box-like sequence, G-X-C-X-G [35], where X is considered to be an arbitrary amino acid, which is actually named after the lipase box sequence (G-X-SX-G) observed in lipases and also in other related enzymes such as PHA depolymerases [36]. The presence of Cys in the lipase box-like sequence is considered to be an active centre of PHA synthases [37].
Amino acid sequence of Bacillus sp.CM27 showed the presence of PhaC box with sequence, G-Y-C-M-G-G having cysteine in the box. The amino acid sequence alignment around the catalytic cysteine from various PHA synthases mainly of Bacillus sp. is shown in Fig. 4. The presence of PhaC box is obvious in all the sequences. There are reports where the PhaCs from Halomonas strains have a similar sequence (S-X-C-X-G) at the active site with the slight difference is the first amino acid that is Gly in the lipase box-like sequence is replaced with Ser [38]. PhaCs from Paracoccus denitrificans and numerous other strains have been projected to have a different active site sequence (G-X-C-X-A) where the Ala is seen to replace second Gly in the sequence [39, 40]. Nambu and coworkers [41] suggested from his extensive work that the consensus sequence [GS]-X-C-X-[GA]-G observed in all characterized PhaCs can be called the ‘PhaC.
In FTIR spectroscopy, the characteristics of PHA band reported by Hong and coworkers [42] were between 1728 and 1744 cm−1 . Misra and coworkers [43] determined key PHA bands at 1724 cm−1 which was similar to PHB standard band obtained (Fig. 5.a) in the current study. Extracted polymer from CM27 grown in Marine basal medium gave PHA band peak at 1733.12 cm−1. Depending on the crystallinity of the PHA and polymer chain length, the peak location of the band is known to differ [42].
Conclusion
This study exploits new insight for unexplored bacterial species from microplastics paving opportunity to find potential bacteria that would produce PHA on an industrial scale. Each bacterial strain responds differently to media conditions, the ratio between carbon and nitrogen of media is very important in relation to both bacterial growth and the efficiency of PHA biosynthesis. Lack of customized knowledge of metabolic pathways limits its complete utilization as PHA producers at optimization stage. Further studies can be carried out by using cheap renewable waste as carbon source to reduce the cost of PHA production, a step ahead in commercialization of PHA bioplastic.
Data Availability
The data that support the findings of the study are available on request froth the corresponding author.
References
Iyer M, Tiwari S, Renu K, Pasha MY, Pandit S, Singh B, Vellingiri B (2021) Environmental survival of SARS-CoV-2–a solid waste perspective. Environ Res 197:111015. https://doi.org/10.1016/j.envres.2021.111015
Amaral-Zettler LA, Zettler ER, Slikas B, Boyd GD, Melvin DW, Morrall CE, Mincer TJ (2015) The biogeography of the Plastisphere: implications for policy. Front Ecol Environ 13(10):541–546. https://doi.org/10.1890/150017
Awasthi AK, Tan Q, Li J (2020) Biotechnological potential for microplastic waste. Trends Biotechnol 38(11):1196–1199. https://doi.org/10.1016/j.tibtech.2020.03.002
Philip S, Keshavarz T, Roy I (2007) Polyhydroxyalkanoates: biodegradable polymers with a range of applications. J chem technol biotechnol 82(3):233–247. https://doi.org/10.1002/jctb.1667
Steinbüchel A, Schlegel HG (1991) Physiology and molecular genetics of poly (β-hydroxyalkanoic acid) synthesis in Alcaligenes eutrophus. Mol Microbiol 5(3):535–542. https://doi.org/10.1111/j.1365-2958.1991.tb00725.x
Kim YB, Lenz RW (2001) Polyesters from microorganisms. Biopolyesters. https://doi.org/10.1007/3-540-40021-4_2
Elain A, Le Fellic M, Corre YM, Le Grand A, Le Tilly V, Audic JL, Bruzaud S (2015) Rapid and qualitative fluorescence-based method for the assessment of PHA production in marine bacteria during batch culture. World J Microbiol Biotechnol 31(10):1555–1563. https://doi.org/10.1007/s11274-015-1904-4
Zettler ER, Mincer TJ, Amaral-Zettler LA (2013) Life in the “plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol 47(13):7137–7146. https://doi.org/10.1021/es401288x
Thompson RC, Olsen Y, Mitchell RP, Davis A, Rowland SJ, John AW, Russell AE (2004) Lost at sea: where is all the plastic? Science 304(5672):838. https://doi.org/10.1126/science.1094559
Auta HS, Emenike CU, Jayanthi B, Fauziah SH (2018) Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar Pollut Bull 127:15–21. https://doi.org/10.1016/j.marpolbul.2017.11.036
Auta HS, Emenike CU, Fauziah SH (2017) Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ Pollut 231:1552–1559. https://doi.org/10.1016/j.envpol.2017.09.043
Yang Y, Liu W, Zhang Z, Grossart HP, Gadd GM (2020) Microplastics provide new microbial niches in aquatic environments. Appl Microbiol Biotechnol 104:6501–6511. https://doi.org/10.1007/s00253-020-10704-x
Morohoshi T, Ogata K, Okura T, Sato S (2018) Molecular characterization of the bacterial community in biofilms for degradation of poly (3-hydroxybutyrate-co-3-hydroxyhexanoate) films in seawater. Microbes Environ 33(1):19–25. https://doi.org/10.1264/jsme2.ME17052
Feng L, He L, Jiang S, Chen J, Zhou C, Qian ZJ, Li C (2020) Investigating the composition and distribution of microplastics surface biofilms in coral areas. Chemosphere 252:126565. https://doi.org/10.1016/j.chemosphere.2020.126565
Chien CC, Chen CC, Choi MH, Kung SS, Wei YH (2007) Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J Biotechnol 132(3):259–263. https://doi.org/10.1016/j.jbiotec.2007.03.002
Liu M, González JE, Willis LB, Walker GC (1998) A novel screening method for isolating exopolysaccharide-deficient mutants. Appl Environ Microbiol 64(11):4600–4602. https://doi.org/10.1128/AEM.64.11.4600-4602.1998
Rawte T, Padte M, Mavinkurve S (2002) Incidence of marine and mangrove bacteria accumulating polyhydroxyalkanoates on the mid-west coast of India. World J Microbiol Biotechnol 18(7):655–659. https://doi.org/10.1023/A:1016872631403
Law JH, Slepecky RA (1961) Assay of poly-β-hydroxybutyric acid. J Bacteriol 82(1):33–36. https://doi.org/10.1128/jb.82.1.33-36.1961
Shamala TR, Chandrashekar A, Vijayendra SVN, Kshama L (2003) Identification of polyhydroxyalkanoate (PHA)-producing Bacillus spp using the polymerase chain reaction (PCR). J Appl Microbiol 94(3):369–374. https://doi.org/10.1046/j.1365-2672.2003.01838.x
Khiyami MA, Al-Fadual SM, Bahklia AH (2011) Polyhydroxyalkanoates production via Bacillus plastic composite support (PCS) biofilm and date palm syrup. J Med Plants Res 5(14):3312–3320. https://doi.org/10.5897/JMPR.9001026
De Smet MJ, Eggink G, Witholt B, Kingma J, Wynberg H (1983) Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol 154(2):870–878. https://doi.org/10.1128/jb.154.2.870-878.1983
Mohammed S, Panda AN, Ray L (2019) An investigation for recovery of polyhydroxyalkanoates (PHA) from Bacillus sp. BPPI-14 and Bacillus sp. BPPI-19 isolated from plastic waste landfill. Int J Biol Macromol 134:1085–1096. https://doi.org/10.1016/j.ijbiomac.2019.05.155
Sreekanth MS, Vijayendra SVN, Joshi GJ, Shamala TR (2013) Effect of carbon and nitrogen sources on simultaneous production of α-amylase and green food packaging polymer by Bacillus sp. CFR 67. J Food Sci Technol 50(2):404–408. https://doi.org/10.1007/s13197-012-0639-6
Salgaonkar BB, Mani K, Braganca JM (2013) Characterization of polyhydroxyalkanoates accumulated by a moderately halophilic salt pan isolate Bacillus megaterium strain H 16. J Appl Microbiol 114(5):1347–1356. https://doi.org/10.1111/jam.12135
Wu Q, Huang H, Hu G, Chen J, Ho KP, Chen GQ (2001) Production of poly-3-hydroxybutyrate by Bacillus sp. JMa5 cultivated in molasses media. Antonie van Leeuwenhoek 80(2):111–118. https://doi.org/10.1023/A:1012222625201
Yüksekdağ ZN, Aslim B, Beyatli Y, Mercan N (2004) Effect of carbon and nitrogen sources and incubation times on poly-beta-hydroxybutyrate (PHB) synthesis by Bacillus subtilis 25 and Bacillus megaterium 12. Afr J Biotech 3(1):63–66. https://doi.org/10.5897/AJB2004.000-2011
Wilkinson JF, Munro ALS (1967) The influence of growth limiting conditions on the synthesis of possible carbon and energy storage polymers in Bacillus megaterium. Microbial physiology and continuous culture. HMSO, London, pp 173–185
Ciesielski S, Możejko J, Przybyłek G (2010) The influence of nitrogen limitation on mcl-PHA synthesis by two newly isolated strains of Pseudomonas sp. J Ind Microbiol Biotechnol 37(5):511–520. https://doi.org/10.1007/s10295-010-0698-5
Kim GJ, Lee IY, Yoon SC, Shin YC, Park YH (1997) Enhanced yield and a high production of medium-chain-length poly (3-hydroxyalkanoates) in a two-step fed-batch cultivation of Pseudomonas putida by combined use of glucose and octanoate. Enzyme Microb Technol 20(7):500–505. https://doi.org/10.1016/S0141-0229(96)00179-2
Lee SY, Wong HH, Choi JI, Lee SH, Lee SC, Han CS (2000) Production of medium-chain-length polyhydroxyalkanoates by high-cell-density cultivation of Pseudomonas putida under phosphorus limitation. Biotechnol Bioeng 68(4):466–470. https://doi.org/10.1002/(SICI)1097-0290(20000520)68:4%3c466::AID-BIT12%3e3.0.CO;2-T
Carnicero D, Fernández-Valverde M, Cañedo LM, Schleissner C, Luengo JM (1997) Octanoic acid uptake in Pseudomonas putida U. FEMS Microbiol Lett 149(1):51–58. https://doi.org/10.1111/j.1574-6968.1997.tb10307.x
Durner R, Zinn M, Witholt B, Egli T (2001) Accumulation of poly [(R)-3-hydroxyalkanoates] in Pseudomonas oleovorans during growth in batch and chemostat culture with different carbon sources. Biotechnol Bioeng 72(3):278–288. https://doi.org/10.1002/1097-0290(20010205)72:3%3c278::AID-BIT4%3e3.0.CO;2-G
Sun Z, Ramsay JA, Guay M, Ramsay BA (2006) Automated feeding strategies for high-cell-density fed-batch cultivation of Pseudomonas putida KT2440. Appl Microbiol Biotechnol 71(4):423–431. https://doi.org/10.1007/s00253-005-0191-7
Kranz RG, Gabbert KK, Locke TA, Madigan MT (1997) Polyhydroxyalkanoate production in Rhodobacter capsulatus: genes, mutants, expression, and physiology. Appl Environ Microbiol 63(8):3003–3009. https://doi.org/10.1128/aem.63.8.3003-3009.1997
Rehm BH (2007) Biogenesis of microbial polyhydroxyalkanoate granules: a platform technology for the production of tailor-made bioparticles. Curr Issues Intest Microbiol 9:41–62
Knoll M, Hamm TM, Wagner F, Martinez V, Pleiss J (2009) The PHA depolymerase engineering database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinform 10:89. https://doi.org/10.1186/1471-2105-10-89
Wittenborn EC, Jost M, Wei Y, Stubbe J, Drennan CL (2016) Structure of the catalytic domain of the class I polyhydroxybutyrate synthase from Cupriavidus necator. J Biol Chem 291:25264–25277. https://doi.org/10.1074/jbc.M116.756833
Ilham M, Nakanomori S, Kihara T, Hokamura A, Matsusaki H, Tsuge T, Mizuno K (2014) Characterization of polyhydroxyalkanoate synthases from Halomonas sp. O-1 and Halomonas elongate DSM2581: site-directed mutagenesis and recombinant expression. Polym Degrad Stabili 109:416–423. https://doi.org/10.1016/j.polymdegradstab.2014.04.024
Ueda S, Yabutani T, Maehara A, Yamane T (1996) Molecular analysis of the poly (3-hydroxyalkanoate) synthase gene from a methylotrophic bacterium, Paracoccus denitrificans. J Bacteriol 178:774–779. https://doi.org/10.1128/jb.178.3.774-779.1996
Rehm BH, Steinbüchel A (1999) Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol 25:3–19. https://doi.org/10.1016/S0141-8130(99)00010-0
Nambu Y, Ishii-Hyakutake M, Harada K, Mizuno S, Tsuge T (2020) Expanded amino acid sequence of the PhaC box in the active center of polyhydroxyalkanoate synthases. FEBS Lett 594(4):710–716. https://doi.org/10.1002/1873-3468.13651
Hong K, Sun S, Tian W, Chen GQ, Huang W (1999) A rapid method for detecting bacterial polyhydroxyalkanoates in intact cells by Fourier transform infrared spectroscopy. Appl Microbiol Biotechnol 51(4):523–526. https://doi.org/10.1007/s002530051427
Misra AK, Thakur MS, Srinivas P, Karanth NG (2000) Screening of poly-β-hydroxybutyrate-producing microorganisms using Fourier transform infrared spectroscopy. Biotech Lett 22(15):1217–1219. https://doi.org/10.1023/A:1005602911977
Acknowledgements
The authors are thankful to Director, HOD of Biological Oceanography Division for providing Laboratory facilities. The authors would like to thank Dr. Mahua Saha and Manish Kumar for helping in Sample collection and thank Dr. Supriya Tilvi and Safia Khan for providing FTIR facility. This work was supported by the funds provided by Department of Biotechnology, Govt. of India, New Delhi, under DBT-JRF programme and Institute projects bearing project code OLP 2005 and GAP 3129. This is NIO Contribution No. 6937.
Funding
The authors would like to thank the Department of Biotechnology, Govt. of India, New Delhi, for providing fellowship and CSIR- National Institute of Oceanography, for financial support from GAP 3129 and OLP 2005 projects.
Author information
Authors and Affiliations
Contributions
HK conceived the study. RK guided the experiments which were carried out. HK and RK searched database for articles and wrote the manuscript. RK analysed DNA sequence of PHA synthase gene. RK critically revised the manuscript. The authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kankonkar, H.T., Khandeparker, R.S. Microplastics a Novel Substratum for Polyhydroxyalkanoate (PHA)-Producing Bacteria in Aquatic Environments. Curr Microbiol 79, 258 (2022). https://doi.org/10.1007/s00284-022-02929-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02929-y