Abstract
Biotic and abiotic stresses are severely limiting plant production and productivity. Of notable importance is salt stress that not only limits plant growth and survival, but affects the soil fertility and threatens agricultural ecosystems sustainability. The problem is exacerbated in fragile arid and semi-arid areas where high evaporation, low precipitation and the use of salty water for irrigation is accelerating soil salinization. Legumes, considered very nutritious foods for people and providing essential nutrients for ecosystems are a fundamental element of sustainable agriculture. They can restore soil health by their ability to fix nitrogen in a symbiotic interaction with the rhizobia of the soil. However, salt stress is severely limiting productivity and nitrogen fixation ability in legumes. Plant growth-promoting rhizobacteria (PGPR) and mainly actinobacteria promote plant growth by producing phytohormones, siderophores, antibiotics and antifungal compounds, solubilizing phosphate and providing antagonism to phytopathogenic microorganisms. In addition, actinobacteria have beneficial effects on nodulation and growth of legumes. In this study, actinobacteria isolated from different niches and having PGP activities were used in co-inoculation experiments with rhizobia in Medicago sativa plants rhizosphere submitted to salt stress. The results indicate that drought- and salinity-tolerant Actinobacteria with multiple PGP traits can potentially increase alfalfa growth under saline conditions, in the presence or absence of symbiotic rhizobial bacteria. Actinobacteria discovered in this study can, therefore, be suitable biofertilizers in the formulation of agricultural products improving plant development, health and productivity in saline soils, a necessary alternative for modern agriculture and sustainable development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adverse conditions due to biotic and abiotic stresses are the main factors limiting agricultural production and productivity [1,2,3,4]. Salinity is a major factor affecting soil fertility and limiting the growth and survival of plants in various parts of the world, particularly in arid and semi-arid areas where high evaporation and low precipitation make irrigation necessary to meet plant water needs [5]. About one-third of irrigated land is affected by salinity [4], mainly because of high temperatures, lack of fresh water and/or poor quality, salt-rich irrigation water [3] and poor management of irrigation [6]. Thus, salt accumulation in soils and groundwater has threatened productivity and soil sustainability [7] and the adverse effect of salinity on plant growth is well established [3]. Plant responses to NaCl stress include a series of changes at the molecular, biochemical and physiological levels, causing a disturbance of the homeostasis and the distribution of ions in the cell and denaturation of structural and functional proteins [4, 8]. Plants often face rapid fluctuations and adversity of environmental conditions due to their intrinsic metabolic abilities [9]. Plants also have the potential to reduce the impact of environmental stresses through the microbiome they harbour [4, 10]. The microbiota provides plants fundamental support for nutrient acquisition, disease resistance and abiotic stress tolerance [4, 11]. Its interaction with plants evokes various types of local and systemic responses that improve the metabolic capacity of plants to fight abiotic stresses [4, 10,11,12].
Legumes are a fundamental element of sustainable agriculture and can offer many economic and environmental benefits. They are very nutritious foods for people and essential nutrients for ecosystems [4]. The integration of legumes into agricultural techniques, such as intercropping, cover crops and crop rotation can restore soil health by their ability to fix nitrogen in a symbiotic interaction with the rhizobia of the soil. Based on their ability to grow on soils low in nitrogen, they can be used effectively to improve saline soil fertility and contribute to the reintroduction of agriculture on these lands [13]. In this line, Sinorhizobium meliloti (strain 1021) and its symbiotic interaction with Medicago sativa is a widely recognized model system for studies of symbiosis and nitrogen fixation. However, salt stress imposes a significant limitation of productivity in legumes. Salinity affects the infection process by inhibiting the growth of absorbent hairs, by decreasing the number of nodules per plant and the amount of nitrogen fixed per unit weight of nodules. Thus, in saline soils, legume yield is reduced due to unsuccessful symbiosis [14].
Plant growth-promoting rhizobacteria (PGPR) are a group of rhizosphere-colonizing bacteria that enhance plant growth, increase yield, improve soil fertility and reduce pathogens and biotic or abiotic stresses. PGPRs help plants by producing growth phytohormones, solubilizing phosphate and by providing antagonism to phytopathogenic microorganisms by producing siderophores, antibiotics and antifungal compounds [4, 10,11,12]. Actinobacteria are widely present in the rhizosphere of plants and produce various agro-active compounds. In the recent years, this group of bacteria, based on its high antimicrobial potential and its dominant saprophytic nature in the soil, has attracted much attention [15]. Actinobacteria promote plant growth by producing phytohormones (Indole Acetic Acid; IAA), siderophores and solubilizing phosphate. Some genera, such Streptomyces exert an immense biocontrol effect on various phytopathogens [14]. By these intrinsic abilities, they colonize successfully plants root systems by releasing many hydrolytic enzymes and can survive in hostile conditions by forming spores. Actinobacteria have long been considered as free-living soil inhabitants, but recently the importance of their complex interactions with plants and other organisms is being discovered and widely documented [16].
Actinobacteria have beneficial effects on nodulation and growth of legumes. Tokala et al. [17] found that the soil isolates Streptomyces lydicus WYEC108 increased the number of nodules, height and weight of pea (Pisum sativum L.) plants. Similarly, co-inoculation of soybean with Streptomyces sp. [18] or Nocardia sp. [19] and Bradyrhizobium japonicum improved the growth of soybean plants. Specific studies on alfalfa have shown growth enhancement associated with an application of Micromonospora spp. [20] alone or in co-inoculation with Sinorhizobium meliloti strain 1021. Solans et al. [21] found that actinobacteria other than Frankia isolated on the surface of the root nodules of Discaria trinervis improved the nodulation of alfalfa in the presence of high levels of nitrogen in the soil which usually inhibit nodulation. A comparative study of total bacterial profiles in soil with those specific for actinobacteria indicates an abundance of actinobacteria in the rhizosphere of legumes [22]. Subsequent studies have shown the beneficial effects of these isolates on legume growth and generated interest in their potential use as co-inoculants with rhizobia in legume crops [22].
Our primary results documented that multiple PGP traits of the salinity-tolerant actinobacteria, can increase alfalfa growth under saline conditions, in the presence or absence of symbiotic rhizobial bacteria. This finding highlights their possible use as biofertilizers for improvement of plant development, health and productivity in saline soils [23]. Therefore, this study aimed to enhance the colonization of the rhizosphere of Medicago sativa host plant by strains of Sinorhizobium meliloti under salt stress after co-inoculation with symbiotic and non-symbiotic actinobacteria isolated from different niches and having PGP activities. The Actinobacterial effect is checked through successful establishment of nitrogen-fixing symbiosis, particularly the formation of nodules and by the morphological and biochemical variations of the host plant.
Materials and Methods
Rhizobial Strains
Rhizobial strains Sinorhizobium meliloti 1021 (R1) and S. meliloti 2011(R2) have been obtained from Prof. Eric Boncompagni (University of Nice Sophia Antipolis, France) [24] and used in this study. Rhizobial strains have been cultured on mannitol yeast extract agar media and incubated for optimal growth at 28 ± 2 °C for 48 h. For storage, cultures have been stored at 4 °C on agar media until subsequent use (Table 1) [25].
Sand Sampling
Five samples of sands were obtained from two geographically distinct locations. The sand of “Melghir Sebkha” located in South East Biskra region [34° 17′ 56.6″ N 6° 21′ 54.56″ E. pH 7.56 ± 0.35, electrical conductivity (EC) = 54.2 ± 2.7 mS/cm] representing saline and arid area and the nodules of Alnus glutinosa which grow in the humid area in front of Guerbes Senhadja river of Skikda region (36° 55′ N 7° 16′ E). No permission was required to collect such samples of sands.
Isolation and Cultivation of Actinobacteria Strains
Two methods were used for bacterial isolation; firstly, the bacteria were isolated from the sand of “Melghir Sebkha”. The bacterial isolation was performed by homogenizing 10 g of sand in 90 mL of sterile physiological water and vigorous shaking for 1 h. The resulting supernatant was diluted in tenfold series (10−1 to 10−5) and plated out on two different media, International Streptomyces Program 2 ISP2 medium and Luedemann medium [25] supplemented with cycloheximide (50 µg/mL). Plates were then incubated at 28 ± 2 °C for 2 weeks. Growing colonies with different morphologies were re-streaked on new Petri dishes until obtaining pure bacterial cultures. Twenty colonies have been collected and tested for their PGP activities. Based on the PGP activities of the bacterial collection 4 strains designated MS1, MS2, MS3 and MS4 were selected for further characterization and plant inoculation studies (Table 1).
The second bacterial isolation was from the second sample, the nodules of Alnus glutinosa were harvested from plant roots after the removal of rhizosphere soil, washed in sterile distilled water, surface-sterilized by H2O2 (30% vol/vol) immersion for 10 min and rinsed with sterile distilled water. Each lobe of nodules was incubated in 5 mL of BAP medium and incubated at 28 ± 2 °C for several weeks with daily removal of culture tubes contaminated with fast-growing bacteria [25]. Thirteen bacterial cultures were obtained and re-streaked on ISP2 agar medium for further growth at 28 ± 2 °C for 2 weeks. All bacterial cultures were screened for their PGP activities and strain Ag-1 selected for subsequent characterization and plant inoculation experiments (Table 1). All isolates were maintained on plates for short-term storage and in medium supplemented with 30% glycerol at −80 °C for long-term storage.
Identification of Actinobacteria by 16S-rRNA Gene Amplification, Sequencing and Phylogenetic Analysis
Bacterial genomic DNA was extracted by a CTAB–SDS lysis protocol [1]. PCR amplification of the 16S-rRNA gene was performed using the universal primers 27 F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1496R (5′-CTACGGCTACCTTGTTACGA-3′) [26]. PCR mixture, amplification conditions and PCR products sequencing were conducted according to Prospero et al. [2]. Partial 16S-rRNA sequences of the isolates were compared with available sequences in the National Centre for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) by BLAST search. Actinobacterial 16S-rDNA sequence alignment and phylogenetic analysis were performed using standard procedures detailed in Mlaik et al. [26].
Biochemical Characterization
Bacterial strains phenotypic screening was performed using different carbon sources and evaluated using standard methods. Degradation of sugars and amino acids used in the time course of the study was evaluated on minimal salt medium [10, 11]. To detect putative extracellular enzymes production of selected isolates, 10 µL of each bacterial suspension culture was plated in specific media. Amylase production was performed on starch media, protease production was performed on skim milk media, chitinase was checked on colloidal chitin agar and cellulase production was evaluated on CMC agar [10,11,12]. After incubation at 28 ± 2 °C for 7 days, positive isolates (after adding revealing reagent) were identified by the presence of a clear zone halo around bacterial colonies suggesting enzyme production by the bacterial isolate. Three replicates were performed per experiment.
Stress Tolerance Studies
Several conditions were used to optimize the growth contention for the bacteria strains used in the current study. First, the ability of bacterial strains to tolerate salt stress, hydric stress and different pH values was performed on ISP2 broth for actinobacteria and YEM broth for rhizobia, supplemented with NaCl (0, 200, 400, 600, 800, 1000 and 1200 mM), PEG8000 (10, 20, 40 and 60%) and having different pH values (4, 7, 9 and 11), respectively. The media were inoculated with 100 μL of bacterial cultures and incubated at 28 ± 2 °C for 7 days. Bacterial growth was then determined by measuring the optical density at 600 nm using a spectrophotometer (Spectronic Genesys 20 Visible Spectrophotometer, Setif, Algeria) and plating on solid media with similar results.
The ability of the strains to tolerate different temperatures was performed by incubating the bacterial cultures at 4, 30, 37, 45 and 55 °C for 7 days and optical density measurement at 600 nm using a spectrophotometer (Spectronic Genesys 20, Setif, Algeria). Minimum of three replicates were performed per experiment.
PGP Activities of Actinobacterial Strains
Growth on Nitrogen-Free Medium
Molecular nitrogen fixation was tested by the ability of the bacterial strains to grow on nitrogen-free medium, the DF salt minimal medium [12].
Ammonia (NH3) and Hydrogen Cyanide (HCN) Production
Ammonia production was revealed by the addition of Nessler’s reagent (0.5 mL) giving a yellow-to-brown colour of peptone water inoculated by bacterial cultures and incubated at 28 ± 2 °C for 7 days [12].
The production of hydrogen cyanide (HCN) was performed on nutrient agar supplemented with glycine (4.4 g/L) [12]. The medium was inoculated with the bacterial strains. Whatman paper impregnated with a solution of sodium picrate (5% picric acid and 2% sodium carbonate) was placed inside the lid of the plates and then sealed with parafilm and incubated at 28 ± 2 °C for 7 days. The development of an orange-brown colour on Whatman paper indicated hydrogen cyanide production.
Phosphate Solubilization
The phosphate solubilization ability was tested on Pikovskaya (PVK) medium containing tricalcium phosphate (Ca3HPO4) as the sole source of phosphate. A volume of 10 μL bacterial culture was spotted on the surface of the PVK agar as described by [11]. After incubation at 28 ± 2 °C for 7 days, the diameter of the clear halo around the colony was measured and used to estimate bacterial ability for phosphate solubilization.
Siderophores Production
Siderophore production was tested in Chrome Azurol S (CAS) medium according to [10]. The actinobacterial inoculated King B liquid medium was incubated at 28 ± 2 °C for 72 h and the resulting cultures were centrifuged at 5000 rpm for 30 min. 500 μL of the supernatant was then mixed with 500 μL CAS solution and OD measured at 630 nm after 20 min of incubation. The percentage of siderophores was evaluated using the following formula: St-Se/St × 100, where St was the OD of the CAS solution (intense blue colour, control) and Se was the OD of the test solution (light blue to orange colour depending on the intensity of production).
Indole Acetic Acid (IAA) Production
The production of indole acetic acid (IAA) was tested on Dworkin and Foster (DF) medium supplemented with 1 g/L tryptophan according to Slama et al. [10]. The cultures were then incubated at 28 ± 2 °C for 7 days and a colourimetric assay was performed using the method detailed in Slama et al. [10]. Briefly, the cultures were centrifuged at 5000 rpm for 20 min and 1 mL of supernatant was mixed with 2 mL Salkowski reagent (50 mL perchloric acid and 1 mL 35% FeCl3 0.5 M). The OD was measured at 530 nm. Concentrations of IAA were determined using a calibration curve prepared from an IAA solution in the range 0 to 10−5 M.
Effects of Bacterial Inoculation and Co-inoculation Experiments on the Nodulation of Medicago sativa L. Plants
Plant Materials
Medicago sativa L. seeds were obtained from the “Institut Technique des Grandes Cultures” (I.T.G.C.) of the city of Setif, Algeria. Alfalfa seeds were surface sterilized using successive incubations in ethanol 70% for 30 s, 4% sodium hypochlorite for 3 min followed by 4 successive washings by sterile distilled water [4]. Surface sterilized seeds were then germinated in dark, in Petri dishes amended with 0.8% water agar solution and incubated at 28 °C for 48 h.
Bacterial Inocula Preparation
Actinobacterial and rhizobial strains were grown on liquid ISP2 and YEM media as previously described and incubated at 28 ± 2 °C for 2 to 7 days. Bacterial cultures densities were then calibrated at 108 cells/mL using a specific correlation between OD and bacterial count in the medium [4].
Bacterial Inoculations and Co-inoculations and Plant Growth Conditions
Surface sterilized alfalfa seeds were sown in sterilized internal surface plastic pots (by several washes using ethanol 70%). Pots were then filled with equal quantities of sand that was previously thoroughly washed with sterilized distilled water and autoclaved (121 °C for 1 h during three cycles separated by 24 h). Surface sterilized seeds were sown aseptically at 1 cm depth at a density of three seeds per pot. After 1 week, alfalfa plants were inoculated with 1 mL of each isolate bacterial suspension taken during the exponential growth phase and calibrated at 108 cells/mL [4]. Inoculation and co-inoculations were performed using rhizobial strains R1 and R2 and actinobacterial strains MS1-4 and Ag1 alone or by joint inoculation with one rhizobial and each actinobacterial selected strains (Table 2). Pots were then placed in a growth chamber with 16 h day/8 h night photoperiod and 26/18 °C day/night temperature. Pots were watered twice weekly with a nitrogen-depleted nutritive solution. The experiment had been repeated six times and after 2 months of culture alfalfa plantlets were collected and the presence of nodules recorded.
Bacterial Inoculations and Co-inoculations and Plant Growth Conditions Under Salt Stress
Based on the results obtained in the “Bacterial inoculations and co-inoculations and plant growth conditions” section experiments, three actinobacterial strains MS1-3 having induced nodulation had been selected to evaluate their efficiency in co-inoculation experiments with rhizobial strains under salt stress. Experiments had been conducted as previously described in the section “Bacterial inoculations and co-inoculations and plant growth conditions” except that plants had been grown with and without salt stress. The experimental set-up was explained in Table 3. Briefly, after 1 week of alfalfa plant growth, bacterial inoculation and co-inoculations had been performed using 1 mL of bacterial suspensions of each isolate at a density of 108 cells/mL. Inoculations and co-inoculations had been repeated three times during 3 weeks and the pots were watered two times a week with a nitrogen depleted nutritive solution. Two weeks post-inoculation and co-inoculation pots of the second group (100 mM NaCl) were watered with the 100 mM saline solution once a week during 1 month (4 times).
Monitoring of Morphological Parameters
After 2 months of culture, alfalfa plantlets were collected and their growth evaluated by estimation of root and shoot fresh weight (g), plantlet aerial part height (cm), root length (cm), leaves and nodules numbers.
Estimation of Photosynthetic Chlorophyll Content
Photosynthetic chlorophyll contents have been determined using the procedure described by Rekik et al. [3]. Briefly, 0.5 g of leaf material of each sample were cut in 1 mm2 pieces and homogenised in 10 mL 80% acetone and incubated at −10 °C overnight. Extract was then centrifuged at 14,000 rpm for 5 min and optical density of the supernatant estimated at 663, 645 and 470 mM. Chlorophyll a (Cha), b (Chb) and total chlorophyll a and b contents (Cha+b) and carotenoids expressed in mg/g were evaluated according to the following equations:
Soluble Amino Acids and Proline Content
Amino acids and proline extraction procedures have been detailed in Cherif-Silini et al. [4]. Briefly, 0.5 g plantlet fresh material was harvested in 5 mL methanol:chloroform:water solution 60:25:15 respectively. The samples were then heated in a water bath at 60 °C for 2 h and centrifuged at 10,000 rpm for 10 min. The supernatant was then used to estimate soluble amino acids and proline contents.
For soluble amino acids content, 1 mL of supernatant was added to 1 mL of acetate buffer (pH 4.3) and 1 mL of ninhydrin solution (5% in ethanol). Samples were then vigorously shaken and heated in a water bath at 95 °C for 15 min. OD was then measured at 570 nm and concentration of soluble amino acids obtained using glycine as standard.
Free proline content had been measured according to Cherif-Silini et al. [4]. Briefly, 1 mL of the supernatant described earlier had been added to 4 mL of ninhydrin solution (5% in ethanol), 4 mL of glacial acetic acid and 1 mL of sterile distilled water. The mixture was then heated in a water bath at 90 °C for 45 min and left to settle and reach room temperature. Then, 4 mL toluene had been added to the mixture and OD of the organic phase estimated at 520 nm. Free proline concentration was determined using a calibration curve with known amounts of free proline added.
Statistical Analysis
All the experiments were performed in triplicate, the results expressed as means ± standard deviation (SD) The statistical analysis of the data was conducted using GraphPad prism 8, one-way- and two-way ANOVA was used to identify the variance between different treatments. The results were considered significant when P values were less than 0.05. post hoc Tukey’s HSD test comparison tests were conducted when a significant difference was encountered.
GenBank Accession Numbers
GenBank accession numbers of the bacteria isolates MS-1, MS-2, Ag-1, MS-3 and MS-4 are MK894856, MK894855, MN420819, MN005930 and MK894857, respectively.
Results
Isolation, Morphological and Molecular Characterization of Rhizobial and Actinobacterial Strains
Growth of rhizobial strains S. meliloti R1 and R2 on yeast mannitol agar (YEM) at 28 °C for 48 h revealed small colonies (1 mm in diameter) of rod-shaped cells. Actinobacterial strains selected cultured on ISP2 media at 28 °C after 2 to 7 days of culture showed smooth colonies having an orange- and red-coloured non-diffusible pigments for MS1 and MS-2, respectively. However, colonies of Ag1, MS3 and MS4 had rough aspects and presented aerial mycelia and a white or grey coloured substrate mycelium. The morphological microscopy of these strains was additionally filamentous (Fig. S1). Majority of the rhizobial and actinobacterial strains catabolized numerous carbon sources and produced different enzymes, such as amylase, protease, chitinase and cellulose (Table S1). 16S-rDNA phylogenetic analysis unambiguously documented that MS1 and MS2, MS3 and MS4 had a homology with the genera Arthrobacter, Streptomyces and Umezawaea, respectively, while Ag-1 lied within the genus Nocardiopsis see (Fig. 1).
Physiological Characterization of Rhizobial and Actinobacterial Strains
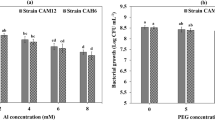
In this study, we had optimised the condition for the bacterial strains, the effect of initial pH and temperature were studied to identify the optimum condition for these bacteria. All strains had been characterized at the physiological level and results were presented in Fig. 2a. Actinobacteria strains were able to grow at pH 7, and can tolerate the extreme pH values. Except MS2 and MS3 were able to tolerate pH values of 9 (Fig. 2a). Among all strains tested for tolerance to different concentrations of PEG rhizobial strains were sensitive to hydric stress. S. meliloti R1 could not grow beyond a 10% PEG concentration compared to S. meliloti R2 strain that could tolerate up to 20% PEG. Actinobacteria on the other hand seemed more adapted to hydric stress mirrored by their ability to grow at high PEG concentrations. MS2 and MS3 for example tolerated up to 60% PEG concentrations (Fig. 2a). All examined rhizobacterial and actinobacterial strains were sensitive to extreme temperatures (4 and 55 °C). Optimal growth temperature for all actinobacterial strains was around 30 °C with strains Ag1 and MS3 was able to grow up to 45 °C (Fig. 2a). Growth of the strains at salt concentrations ranging from 0 to 1200 mM revealed a good ability of MS2 and MS4 to tolerate up to 800 mM salt in the culture medium. NaCl sensitivity was noticed for S. meliloti R1 that did not tolerate salt at any of the salt tested concentrations. S. meliloti R2 and MS1, however, were able to grow up to 400 mM salt concentrations. MS3 was also able to tolerate up to 200 mM salt (Fig. 2a).
a PGP activities and stress tolerance of bacterial and actinobacterial strains used in the study. b PGP activities of actinobacterial strains. The data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at P < 0.05 using Tukey’s HSD test
PGP Activities of Rhizobial and Actinobacterial Strains
All selected actinobacterial strains produced ammonia and efficiently fixed nitrogen, however, none of them was able to produce HCN (Fig. 2a). Siderophore, IAA and phosphate solubilization were detected in all actinobacterial strains but to varying levels. IAA production varied from 7.3 to 31.2 µg/mL with strains MS1 and MS4 being the best producers with 20.15 and 31.18 µg/mL, respectively. Siderophore production reached high levels of activity mainly for strains MS2, MS3 and MS4 producing 63.13, 80.9 and 74.62%, respectively (Fig. 2b). Phosphate solubilization was observed only for two strains MS1 and MS4 with halo diameters reaching 18 and 13 mm, respectively. All other strains had weak phosphate solubilization abilities (Fig. 2b).
Morphological Parameters of Bacterial Plants Inoculation and Co-inoculation Experiments
Variance analysis of root and shoot length, root and shoot fresh weight and leaves and nodule numbers documented that bacterial inoculation was beneficial to alfalfa plants and that salinity severely and significantly interfered with these growth parameters (P < 0.05). In both inoculation and co-inoculation experiments with and without salt stress, morphological parameters of alfalfa plants inoculated with rhizobia alone or co-inoculated with actinobacteria showed a significant effect of co-inoculation of S. meliloti R1 and R2 strains with actinobacterial isolates MS1, MS2 and Ag1 (Fig. 3). Bacterial inoculation experiments and plant growth under salt stress results indicated that salinity affected the shoot part and the root length. However, the treatment with co-inoculation showed a significant increase P < 0.0001 in the shoot (131–256%) and root length (56–135%) in comparison to the negative control. The optimum length was achieved in the inoculant by the plants S. meliloti R1 and R2 strains with actinobacterial isolates MS1, MS2 and Ag1 (Fig. 3a, b). Plant aerial parts and roots fresh weight were significantly improved in alfalfa plants co-inoculated with S. meliloti R1 and R2 strains. The actinobacterial strains MS1, MS2 and Ag1 showed a significant increase of 2 to 3 times the weight of shoots and roots in the absence and in the presence of salt stress as compared to the negative control (Fig. 3c, d). In addition, bacterial co-inoculation experiments improved the numbers of leaves and nodules. The plants co-inoculated showed a number of nodules increased by 61 and 51% for the strains R1 and R2, respectively as compared to the plants inoculated alone (Fig. 3e,f). In experimental conditions with NaCl stress, nodule number was significantly higher with R1 (36%) and R2 (80%) than control inoculations under NaCl stress (Fig. 3f). Nodulation ability of actinobacterial strains in co-inoculation experiments with S. meliloti R1 and R2, clearly documented that three strains MS1, MS2 and Ag1 were able to allow nodule formations on Medicago sativa roots (Fig. 3f).
a, b Root length and shoot length (mm) of Medicago sativa against different treatments of rhizobacterial and actinobacterial strains and NaCl treatments. c, d Root weight and shoot weight (g) of Medicago sativa against different treatments of rhizobacterial and actinobacterial strains and NaCl treatments. e, f Number of leaves and nodules of Medicago sativa against different treatments of rhizobacterial and actinobacterial strains and NaCl treatments. The data present mean ± standard error. Bars labelled with asterisk are significantly different among the treatments at P < 0.05 using ANOVA analysis
Biochemical Parameters of Bacterial Plants Inoculation and Co-inoculation Experiments
Chlorophyll a, b (Fig. 4a, b) and total chlorophyll (Fig. 4c) and carotenoid contents (Fig. 4d) were significantly improved in co-inoculated plants with and without NaCl stress. Proline accumulation and free amino acid levels in leaves of alfalfa plants co-inoculated with rhizobial and actinobacterial strains were significantly lower than control levels. These finding clearly illustrated that actinobacterial strains could mitigate salt stress that was reflected by lower levels of proline and free amino acids in leaves of inoculated and co-inoculated plants (Fig. 5a, b).
a Chlorophyll a, b Chlorophyll b, c Chlorophyll a + b, d Carotenoids (mg/g FW) of Medicago sativa against different treatments of rhizobacterial and actinobacterial strains and NaCl treatments. The data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at P < 0.05 using Tukey’s HSD test
a Amino acids and b proline content (µg/g FW) of Medicago sativa against different treatments of rhizobacterial and actinobacterial strains and NaCl treatments. The data present mean ± standard error. Bars labelled with different letters are significantly different among the treatments at P < 0.05 using Tukey’s HSD test
Discussion
Biological nitrogen fixation plays a crucial role in improving soil productivity [27]. Therefore, nitrogen deficiency severely limits plant growth and productivity [5]. Rhizobium Legume Symbiosis is an economically sound nitrogen source providing necessary nitrogen amounts for efficient plant growth and development and an efficient sustainable agricultural practice as compared to external fertilizer supplies that are costly for the farmers and damaging to the environment [28, 29]. Unfortunately, numerous environmental factors limit nitrogen fixation affecting survival and rhizobia and the rate of infectivity [30, 31]. Recent trends in the field aiming the development of stress-tolerant crops suggest the use of plant growth-promoting (PGP) bacteria as an efficient mean to improve biological nitrogen fixation [32].
In the current study, three selected bacterial strains affiliated to the Actinobacteria phylum were able to enhance nitrogen-fixing symbiosis between S. meliloti and the legume M. sativa. The promotion of nitrogen-fixing symbiosis by actinobacteria has been widely documented [33]. Recent studies focus more on PGP potential of actinobacteria as compared to other bacteria, due to their relative abundance in the soil and their ability to produce a variety of metabolites. The selected strains showed these abilities, they can degrade various carbonaceous substances. These characteristics allowed not only to study their metabolic profile but also to understand their ability to compete and survive [34].
Also, it has been reported that a large part of their genome (~5–10%) is devoted to the production of secondary metabolites which help them to grow even in extreme conditions. Actinobacteria are widespread in aquatic and terrestrial habitats, including extreme habitats and hyper-arid desert soils [13]. This was clearly shown by the tolerance of these strains to abiotic stress, particularly aridity and salinity. Given the origin of their isolation, some strains can tolerate salt concentrations up to 800 mM, an alkaline pH 9, a temperature of 45 °C and a PEG osmotic pressure of 60%. Soil characteristics play a prominent role in the microbial selection process. Bacteria isolated from extreme environments can survive inhibitory parameters compared to those isolated from non-stressful habitats [35].
It is well documented that Actinobacteria, like other PGPRs, employ direct and indirect mechanics to boost crops growth and protection against diseases. In the present study, all our isolates produced IAA at variable rates. It is well documented that rhizospheric and endophytic actinobacteria can provide phytohormones to their plant hosts [16, 36]. IAA is the main phytohormone that boosts plant growth and development. IAA, besides being critical for nodule formation, have also been reported as essential trigger of cell proliferation and differentiation and formation. of vascular bundles [37]. As previously reported in previous studies, the phosphate solubilization ability was also present in our isolates [38]. It is worth noticing that in the report of Fernandez et al. [39] phosphate solubilizing bacteria have favourable effects on soybean growth. This improved the nutrient supply of Rhizobium symbionts and subsequent nodules formation. Other PGP traits have been shown in the selected actinobacteria like siderophores production. The genus Streptomyces was well known for its characteristic siderophores types, such as desferrioxamine and coelichelin [40] that not only play a major role in plant nutrition, but also provided plant protection via phytopathogen control. The ability to fix nitrogen in these strains, revealed by strains growth on nitrogen-free medium, has also been reported [41]. It has been documented that PGP actinobacteria can minimize and cope with the adverse effects of biotic and abiotic stresses [14, 42]. Numerous studies suggested that biotic abiotic stresses mitigation in plants by PGP actinobacteria is done through cell wall degrading enzymes (protease, cellulase, chitinase), secondary metabolite production, low molecular weight inhibitors substances (ammonia for example) and nutrients competition. Jog et al. [42] suggested that these modes of actions should be major properties of any effective fertilizer. In the present study, all selected strains showed multiple PGP traits, independently of their isolation origin. Except the Ag1 strain, all other strains were non-rhizospheric or endophytic bacteria and to our knowledge, this is the first study showing that this kind of bacteria can be used as a plant growth promoter.
Our results clearly showed the importance of actinobacterial inoculants in stimulating the growth of M. sativa plants. Improvement in nodulation and nitrogen fixation in alfalfa cultivars was related to the co-inoculation treatment. It was noted that plants co-inoculated with actinobacteria and Sinorhizobium had increased nodulation and plant growth compared to plants with single inoculation. Similar results have been previously reported by Gregor et al. [43] using Streptomyces kanamyceticus and Bradyrhizobium japonicum combination for inoculation of soybean. The authors reported an increased nodulation (55%) and nitrogen composition (41%) of soybean stems. In another study Soe and Yamakawa [44] reported an improved nodulation, nitrogen fixation and seed yield of different soybean varieties after co-inoculation of Bradyrhizobium yuanmingense MAS34 and Streptomyces griseoflavus P4. Moreover, the study of Volpin et al. [45] suggested the evidence of a more direct effect of Azospirillum brasilense on nodulation mediated by increased flavonoids exudation in the rhizosphere of alfalfa and common bean. Flavonoids are believed to be plant signalling molecules essential for the establishment of rhizobia–legume symbiosis. It is widely believed that legume nodules are often occupied by a phylogenetically diverse bacterial microbiome. These bacteria have wide effects on plant growth and health as well as nodule formation and nitrogen fixation. However, their precise ecological roles remain unknown [46]. A non-passive role of the non-rhizobial bacteria in nodules has been suggested including manipulation of the plant host [47]. Liu et al. [20] proved that some non-rhizobial bacteria improve the nodulation and nitrogen fixation of leguminous symbionts–rhizobia and help Rhizobium extending its host range. They selectively control the entry of bacteria in this specific niche, the root nodule and allow their survival [48]. Molecular studies have shown that many actinobacteria can occur as endophytes in various leguminous plants [11]. Streptomyces inoculants application to enhance plant nodulation and increase soil nitrogen content indicated their potent effect on rhizobia–legume symbiosis non-specifically [26]. However, in our case, Streptomyces did not seem to contribute to nodulation as compared to the genus Arthrobacter which not only improved nodulation, but also all the plant growth parameters, in the presence of S. meliloti R1 or R2.
Variance analysis showed that alfalfa growth parameters were negatively affected by salinity. Bacterial inoculation has been reported to reduce the undesirable effects of stress on plant growth [4]. Our results were in agreement with earlier findings that inoculated plants grew better and had higher biomass than non-inoculated plants under salt stress conditions. Legume–rhizobia symbiosis and the process of initiation of nodules on legumes were both sensitive to salt stress [49]. Salt stress inhibited the early stages of rhizobia–legume symbiosis. The adverse effects of salinity on symbiosis may also result from the suppression of nodule function and the reduction of plants’ ability to grow [49]. The increased growth of plants co-inoculated with non-rhizobial strains as compared to plants inoculated with the rhizobial strain S. meliloti alone can be attributed to the higher tolerance of actinobacteria strains to salinity and drought and to their greater ability to produce IAA hormone (MS1 and MS2).
Saline stress had also deleterious effect on biochemical parameters resulting in a decrease in chlorophyll concentration [50] due to the weakening of protein–lipid complexes [50] and consequently to the reduction in interception of light, an increase in the activity of chlorophyllase and the synthesis of nitrogen compounds that consumes a large quantity of nitrogen [51]. It was also known that higher concentrations of ions in saline soils accumulate in plant cells, inactivate enzymes and inhibit protein synthesis and photosynthesis [3].
Analysis of the leaf proline content revealed that salinity stimulated its intracellular synthesis. This synthesis was at the maximum level at 100 mM NaCl. Inoculation induced a decrease in leaf content under the effect of salt stress. Proline is the amino acid most widely accumulated by plants in saline soil [4]. Inoculated plants can improve their growth rate and salt tolerance and suppress the harmful effects of stress, allowing them to adapt to salt conditions [4]. The results of this study indicate that drought- and salinity-tolerant actinobacteria with multiple PGP traits can potentially increase alfalfa growth under saline conditions, in the presence or absence of symbiotic rhizobial bacteria. This knowledge will be useful to define strategies for the application of these bacteria as bio-inoculants, alone or associated with rhizobial bacteria. Such approaches will improve the performance or persistence of rhizobia and reduce the use of chemical fertilizers [28]. According to the results of this study, MS1 and MS2 bacteria can be suitable biofertilizers in the formulation of agricultural products improving plant development, health and productivity in saline soils, a necessary alternative for modern agriculture and sustainable development.
Conclusions
Our study highlights the potential use of Actinobacteria and mainly those discovered in this study as biofertilizers in the formulation of agricultural products improving plant development, health and productivity in saline soils, a necessary alternative for modern agriculture and sustainable development.
References
Luchi N, Ghelardini L, Belbahri L, Quartier M, Santini A (2013) Rapid detection of Ceratocystis platani inoculum by quantitative real-time PCR assay. Appl Environ Microbiol 79(17):5394–5404. https://doi.org/10.1128/AEM.01484-13
Prospero S, Vercauteren A, Heungens K, Belbahri L, Rigling D (2013) Phytophthora diversity and the population structure of Phytophthora ramorum in Swiss ornamental nurseries. Plant Pathol 62:1063–1071. https://doi.org/10.1111/ppa.12027
Rekik I, Chaabane Z, Missaoui A, Chenari Bouket A, Luptakova L, Elleuch A, Belbahri L (2017) Effects of untreated and treated wastewater at the morphological, physiological and biochemical levels on seed germination and development of sorghum (Sorghum bicolor (L.) Moench), alfalfa (Medicago sativa L.) and fescue (Festuca arundinacea Schreb.). J Hazard Mater 326:165–176
Cherif-Silini H, Thissera B, Bouket AC, Saadaoui N, Silini A, Eshelli M, Alenezi FN, Vallat A, Luptakova L, Yahiaoui B, Cherrad S, Vacher S, Rateb ME, Belbahri L (2019) Durum wheat stress tolerance induced by endophyte Pantoea agglomerans with genes contributing to plant functions and secondary metabolite arsenal. Int J Mol Sci 20:3989
Wang C, Wu J, Zeng W, Zhu Y, Huang J (2019) Five-year experimental study on effectiveness and sustainability of a dry drainage system for controlling soil salinity. Water 11(1):111. https://doi.org/10.3390/w11010111
Plaut J, Wadsworth WD, Pangle R, Yepez EA, McDowell NG, Pockman WT (2013) Reduced transpiration response to precipitation pulses precedes mortality in a piñon–juniper woodland subject to prolonged drought. New Phytol 200(2):375–387. https://doi.org/10.1111/nph.12392
Daliakopoulos IN, Tsanis IK, Koutroulis A, Kourgialas NN, Varouchakis AE, Karatzas GP, Ritsema CJ (2016) The threat of soil salinity: a European scale review. Sci Total Environ 573:727–739. https://doi.org/10.1016/j.scitotenv.2016.08.177
Hashem A, Abd-Allah EF, Alqarawi A, Al-Huqail AA, Wirth S, Egamberdieva D (2016) The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front Plant Sci 7:1089. https://doi.org/10.3389/fmicb.2016.01089
Simontacchi M, Galatro A, Ramos-Artuso F, Santa-María GE (2013) Plant survival in a changing environment: the role of nitric oxide in plant responses to abiotic stress. Front Plant Sci 6:977. https://doi.org/10.3389/fpls.2015.00977
Slama HB, Cherif-Silini H, Chenari Bouket A, Qader M, Silini A, Yahiaoui B, Alenezi FN, Luptakova L, Triki MA, Vallat A, Oszako T, Rateb ME, Belbahri L (2019) Screening for Fusarium antagonistic bacteria from contrasting niches designated the endophyte Bacillus halotolerans as plant warden against Fusarium. Front Microbiol 9:3236. https://doi.org/10.3389/fmicb.2018.03236
Slama HB, Triki MA, Chenari Bouket A, Mefteh FB, Alenezi FN, Luptakova L, Cherif-Silini H, Vallat A, Oszako T, Gharsallah H, Belbahri L (2019) Screening of the high-rhizosphere competent Limoniastrum monopetalum’ culturable endophyte microbiota allows the recovery of multifaceted and versatile biocontrol agents. Microorganisms. https://doi.org/10.3390/microorganisms7080249
Mefteh FB, Daoud A, Chenari Bouket A, Thissera B, Kadri Y, Cherif-Silini H, Eshelli M, Alenezi F, Vallat A, Oszako T, Kadri A, Ros-Garcia JM, Rateb M, Gharsallah N, Belbahri L (2018) Date palm trees root-derived endophytes as fungal cell factories for diverse bioactive metabolites. Int J Mol Sci 19(7):1986. https://doi.org/10.3390/ijms19071986
Fernandez AL, Fabrizzi KP, Tautges NE, Lamb JA, Sheaffer CC (2017) Cutting management and alfalfa stand age effects on organically grown corn grain yield and soil N availability. Renew Agric Food Syst 34(2):1–11
Mathew BT, Torky Y, Amin A, Mourad A-HI, Ayyash MM, El-Keblawy A, El-Keblawy A, AbuQamar SF, El-Tarabily KA (2020) Halotolerant marine rhizosphere-competent actinobacteria promote Salicornia bigelovii growth and seed production using seawater irrigation. Front Microbiol 11:552
Frikha DD, Zarai Z, Najah S, Abdennabi R, Belbahri L, Rateb ME, Mejdoub H, Maalej S (2017) Antagonistic properties of some halophilic Thermoactinomycetes isolated from superficial sediment of a solar saltern and production of cyclic antimicrobial peptides by the novel isolate Paludifilum halophilum. Biomed Res Int 2017:1205258. https://doi.org/10.1155/2017/1205258
El-Tarabily KA, ElBaghdady KZ, AlKhajeh AS, Ayyash MM, Aljneibi RS, El-Keblawy A, AbuQamar SF (2020) Polyamine-producing actinobacteria enhance biomass production and seed yield in Salicornia bigelovii. Biol Fertil Soils 56:499–519
Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA, Bailey JF, Morra MJ (2002) Novel plant microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68:2161–2171
Soe KM, Bhromsiri A, Karladee D, Yamakawa T (2012) Effects of endophytic actinomycetes and Bradyrhizobium japonicum strains on growth, nodulation, nitrogen fixation and seed weight of different soybean varieties. Soil Sci Plant Nutr 58:319–325. https://doi.org/10.1080/00380768.2012.682044
Nimmnoi P, Pongsilp N, Lumyong S (2014) Co-inoculation of soybean (Glycine max) with act in oomycetes and Bradyrhizobium japonicum enhances plant growth, nitrogenase activity and plant growth. J Plant Nutr 37:432–446
Martinez-Hidalgo P, Galindo-Villardon P, Igual JM, Martinez-Molina E (2014) Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.): a new promising plant probiotic bacteria. Sci Rep 4:1–9
Solans M, Vobis G, Wall LG (2009) Saprophytic actinomycetes promote nodulation in Medicago sativa–Sinorhizobium meliloti symbiosis in the presence of high nitrogen. J Plant Growth Regul 28:106–114
Liu C, Zhuang X, Yu Z, Wang Z, Wang Y, Guo X, Xiang W, Huang S (2019) Community structures and antifungal activity of root-associated endophytic actinobacteria of healthy and diseased soybean. Microorganisms 7:243
Belbahri L, Chenari Bouket A, Rekik I, Alenezi FN, Vallat A, Luptakova L, Petrovova E, Oszako T, Cherrad S, Vacher S, Rateb ME (2017) Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front Microbiol 8:1438. https://doi.org/10.3389/fmicb.2017.01438
Galibert F, Finan TM, Long SR, Puhler A, Abola P et al (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293(5530):668–672
Louati M, Ennis NJ, Ghodhbane-Gtari F, Hezbri K, Sevigny JL, Fahnestock MF, Cherif-Silini H, Bryce JG, Tisa LS, Gtari M (2019) Elucidating the ecological networks in stone-dwelling microbiomes. Environ Microbiol. https://doi.org/10.1111/1462-2920.14700
Mlaik N, Bakonyi J, Borsodi A, Woodward S, Belbahri L, Mechichi T (2015) Microbial diversity in tanning wastewaters treatment reactors. Environ Prog Sustain Energy 34(2):401–410. https://doi.org/10.1002/ep.12000
Thilakarathna MS, Chapagain T, Ghimire B, Pudasaini R, Tamang BB, Gurung K, Choi K, Rai L, Magar S, Bk B, Gaire S, Raizada MN (2019) Evaluating the effectiveness of Rhizobium inoculants and micronutrients as technologies for nepalese common bean smallholder farmers in the real-world context of highly variable hillside environments and indigenous farming practices. Agriculture 9:20
Sachs JL, Quides KW, Wendlandt CE (2019) Legumes versus rhizobia: a model for ongoing conflict in symbiosis. New Phytol 219(4):1199–1206
Cherrad S, Charnay A, Hernandez C, Steva H, Belbahri L, Vacher S (2018) Emergence of boscalid-resistant strains of Erysiphe necator in French vineyards. Microbiol Res 216:79–84. https://doi.org/10.1016/j.micres.2018.08.007
Andrews M, Andrews ME (2019) Specificity in legume–rhizobia symbioses. Int J Mol Sci 18(4):26
Ferguson BJ, Mens C, Hastwell AH, Zhang M, Su H, Jones CH, Chu X, Gresshoff PM (2019) Legume nodulation: the host controls the party. Plant Cell Environ 42(1):41–51
Htwe AZ, Moh SM, Soe KM, Moe K, Yamakawa T (2019) Effects of biofertilizer produced from Bradyrhizobium and Streptomyces griseoflavus on plant growth, nodulation, nitrogen fixation, nutrient uptake, and seed yield of mung bean, cowpea, and soybean. Agronomy 9(2):77. https://doi.org/10.3390/agronomy9020077
Solans M, Ruiz OA, Wall LG (2015) Effect of actinobacteria on Lotus tenuis–Mesorhizobium loti symbiosis: preliminary study. Symbiosis 65:33–37
Goodfellow M, Williams ST (1983) Ecology of actinomycetes. Annu Rev Microbiol 37:189–216. https://doi.org/10.1146/annurev.mi.37.100183.001201
Okoro CK, Brown R, Jones AL et al (2009) Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 95:121. https://doi.org/10.1007/s10482-008-9295-2
El-Tarabily KA, Sivasithamparam K (2006) Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol Biochem 34:1–16
Choudhury SR, Johns SM, Pandey S (2019) A convenient, soil-free method for the production of root nodules in soybean to study the effects of exogenous additives. Plant Direct. https://doi.org/10.1002/pld3.135
Jog R, Pandhya M, Nareshkumar G, Rajkumar S (2014) Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiology 160:778–788
Fernandez LA, Zalba P, Gomez MA, Sagordoy MA (2007) Phosphate-solubilization activity of bacterial strains in soil and their effect on soybean growth under greenhouse conditions. Boil Fertil Soil 43:805–810
Barona-Gomez F, Lautru S, Francou FX, Leblond P, Pernodet JL, Challis GL (2006) Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 152(Pt 11):3355–3366
Sellstedt A, Richau KH (2013) Aspects of nitrogen-fixing actinobacteria, in particular free-living and symbiotic Frankia. FEMS Microbiol Lett 342:179–186
Jog R, Nareshkumar G, Rajkumar S (2016) Enhancing soil health and plant growth promotion by actinomycetes. In: Gopalakrishnan S, Sathya A, Vijayabharathi R (eds) Plant growth promoting actinobacteria. Springer, Singapore, pp 33–45
Gregor AK, Klubek B, Varsa EC (2003) Identification and use of actinomycetes for enhanced nodulation of soybean co-inoculated with Bradyrhizobium japonicum. Can J Microbiol 49:483–491
Soe KM, Yamakawa T (2013) Evaluation of effective Myanmar Bradyrhizobium strains isolated from Myanmar soybean and effects of co-inoculation with Streptomyces griseoflavus P4 for nitrogen fixation. Soil Sci Plant Nutr 59:361–370
Volpin H, Burdman S, Castro-Sowinski S, Kapulnik Y, Okon Y (1996) Inoculation with Azospirillum increased exudation of rhizobial nod-gene inducers by alfalfa roots. Mol Plant–Microbe Interact 9(5):388–394
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42. https://doi.org/10.1080/07352689.2014.897899
Gano-Cohen KA, Stokes PJ, Blanton MA, Wendlandt CE, Hollowell AC, Regus JU, Kim D, Patel S, Pahua VJ, Sachs JL (2016) Non-nodulating Bradyrhizobium spp. modulate the benefits of legume-rhizobium mutualism. Appl Environ Microbiol 82(17):5259–5268. https://doi.org/10.1128/AEM.01116-16
Mus F, Crook MB, Garcia K, Costas AG, Geddes BA, Kouri ED et al (2016) Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol 82:3698–3710. https://doi.org/10.1128/AEM.01055-16
Elsheikh EAE, Wood M (1990) Effect of salinity on growth, nodulation and nitrogen yield of chickpea (Cicer arietinum L.). J Exp Bot 41:1263–1269
Rosa-Ibarra MDL, Maiti RK (1995) Biochemical mechanism in glossy sorghum lines for resistance to salinity stress. J Plant Physiol 146(4):515–519
Levitt J (1980) Chilling, freezing, and high temperature stresses. In: Responses of plants to environmental stress, 2nd eds., vol 1
Acknowledgements
This manuscript received funding from Algerian Ministry of Higher Education and Scientific Research to HCS. This organizations provided finance support for this study, but did was not involved in the study design, data analysis, decision to publish, or in writing this manuscript.
Funding
Open Access funding provided by Université de Neuchâtel.. There is no financial support for this work.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: CHS, AS and LB. Performed the experiments: SS, CHS, AS, LL and LB. Analyzed the data: SS, CHS, ACB, AS, FNA, LLand ME. Contributed reagents/materials/analysis tools: LB, CHS, and AS. Wrote and enriched the literature: LB, CHS, ME, ACB and AS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saidi, S., Cherif-Silini, H., Chenari Bouket, A. et al. Improvement of Medicago sativa Crops Productivity by the Co-inoculation of Sinorhizobium meliloti–Actinobacteria Under Salt Stress. Curr Microbiol 78, 1344–1357 (2021). https://doi.org/10.1007/s00284-021-02394-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02394-z