Abstract
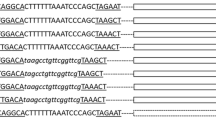
Integrons play important roles in the dissemination of antimicrobial resistant genes among bacteria. Class 2 integrons usually has an internal stop codon, TAA, in integrase genes (intI2), leading to a truncated integrase, IntI2*. However, a few class 2 integrons with a natural full-length integrase have been reported. In this study, the sequences of natural full-length intI2 were extracted from INTEGRALL database and analyzed. A total of 236 sequences of intI2 were retrieved from INTEGRALL database, only seven of which were natural full-length intI2 genes and could be divided into five types according to their coding amino acid sequence. Quantitative real-time PCR was used to detect gene cassette sat2 integration and excision efficiency catalyzed by different natural full-length IntI2s. The results showed that all five IntI2s could catalyze attI2 × attCsat2 integration and attCdfrA1/sat2 × attCsat2/aadA1 excision in Escherichia coli. Integration and excision frequency catalyzed by IntI2A176 was highest and was about twofold as high as those catalyzed by IntI2S175_A176. The secondary structure of the IntI2 was predicted by online software. Polymorphisms of these five IntI2s were limited within residues 172, 174, 175, 176 and 256, and these residues were all far away from the predicted DNA binding regions or catalyzed sites. Influence of amino acid sequence polymorphisms of these natural full-length IntI2s on their catalyzed activities is limited.
Similar content being viewed by others
References
Laxminarayan R, Chaudhury RR (2016) Antibiotic resistance in India: drivers and opportunities for action. PLoS Med 13:e1001974. https://doi.org/10.1371/journal.pmed.1001974
Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R (2014) Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750. https://doi.org/10.1016/S1473-3099(14)70780-7
Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O (2013) Antibiotic resistance-the need for global solutions. Lancet Infect Dis 13:1057–1098. https://doi.org/10.1016/S1473-3099(13)70318-9
Harbottle H, Thakur S, Zhao S, White DG (2006) Genetics of antimicrobial resistance. Anim Biotechnol 17:111–124. https://doi.org/10.1080/10495390600957092
Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart JT, Hossain A, Johnson JA, Goering RV, Thomson KS (2003) Plasmid mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumonia isolates. J Antimicrob Chemother 51:711–714
Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, Medalla F, Chiller TM, Hooper DC (2006) Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis 43:297–304. https://doi.org/10.1086/505397
Stokes HW, Hall RM (1989) A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 3:1669–1683. https://doi.org/10.1111/j.1365-2958.1989.tb00153.x
Mazel D (2006) Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. https://doi.org/10.1038/nrmicro1462
Escudero JA, Loot C, Nivina A, Mazel D (2015) The integron: adaptation on demand. Microbiol Spectr 3:MDNA3-0019–2014. https://doi.org/10.1128/microbiolspec.MDNA3-0019-2014
Gillings MR (2014) Integrons: past, present, and future. Microbiol Mol Biol Rev 78:257–277. https://doi.org/10.1128/MMBR.00056-13
Fluit AC, Schmitz FJ (1999) Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur J Clin Microbiol Infect Dis 18:761–770. https://doi.org/10.1007/s100960050398
Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM (1993) Site-specific insertion of gene cassettes into integrons. Mol Microbiol 9:41–52. https://doi.org/10.1111/j.1365-2958.1993.tb01667.x
Collis CM, Hall RM (1992) Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol 174:1574–1585. https://doi.org/10.1128/jb.174.5.1574-1585.1992
Gravel A, Fournier B, Roy PH (1998) DNA complexes obtained with the integron integrase IntI1 at the attI1 site. Nucleic Acids Res 26:4347–4355. https://doi.org/10.1093/nar/26.19.4347
Recchia GD, Stokes HW, Hall RM (1994) Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res 22:2071–2078. https://doi.org/10.1093/nar/22.11.2071
Collis CM, Hall RM (1995) Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 39:155–162. https://doi.org/10.1128/aac.39.1.155
Jove T, Da RS, Denis F, Mazel D, Ploy MC (2010) Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6:e1000793. https://doi.org/10.1371/journal.pgen.1000793
Papagiannitsis CC, Tzouvelekis LS, Miriagou V (2009) Relative strength of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrob Agents Chemother 53:277–280
Wei Q, Jiang X, Li M, Chen X, Li G, Li R, Lu Y (2011) Transcription of integron-harboured gene cassette impacts integration efficiency in class 1 integron. Mol Microbiol 80:1326–1336. https://doi.org/10.1111/j.1365-2958.2011.07648.x
Hansson K, Sundstrom L, Pelletier A, Roy PH (2002) IntI2 integron integrase in Tn7. J Bacteriol 184:1712–1721. https://doi.org/10.1128/jb.184.6.1712-1721.2002
Ramirez MS, Quiroga C, Centron D (2005) Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 49:5179–5181. https://doi.org/10.1128/AAC.49.12.5179-5181.2005
Marquez C, Labbate M, Ingold AJ, Roy CP, Ramirez MS, Centron D, Borthagaray G, Stokes HW (2008) Recovery of a functional class 2 integron from an Escherichia coli strain mediating a urinary tract infection. Antimicrob Agents Chemother 52:4153–4154. https://doi.org/10.1128/AAC.00710-08
Wei Q, Hu Q, Li S, Lu H, Chen G, Shen B, Zhang P, Zhou Y (2014) A novel functional class 2 integron in clinical Proteus mirabilis isolates. J Antimicrob Chemother 69:973–976. https://doi.org/10.1093/jac/dkt456
Barlow RS, Gobius KS (2006) Diverse class 2 integrons in bacteria from beef cattle sources. J Antimicrob Chemother 58:1133–1138. https://doi.org/10.1093/jac/dkl423
Rodriguez-Minguela CM, Apajalahti JH, Chai B, Cole JR, Tiedje JM (2009) Worldwide prevalence of class 2 integrases outside the clinical setting is associated with human impact. Appl Environ Microbiol 75:5100–5110. https://doi.org/10.1128/AEM.00133-09
Jove T, Da RS, Tabesse A, Gassama-Sow A, Ploy MC (2017) Gene expression in class 2 integrons is SOS-independent and involves two Pc promoters. Front Microbiol 8:1499. https://doi.org/10.3389/fmicb.2017.01499
Mendes MA, Couve-Deacon E, Bousquet P, Chainier D, Jove T, Ploy MC, Barraud O (2019) Proteae: a reservoir of class 2 integrons? J Antimicrob Chemother 74:1560–1562. https://doi.org/10.1093/jac/dkz079
Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A (2009) INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. https://doi.org/10.1093/bioinformatics/btp105
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Yang Z, Jiang X, Wei Q, Chen N, Lu Y (2009) A novel and rapid method for determining integration frequency catalyzed by integron integrase intI1. J Microbiol Methods 76:97–100. https://doi.org/10.1016/j.mimet.2008.09.002
MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN (2006) Structural basis for broad DNA-specificity in integron recombination. Nature 440:1157–1162. https://doi.org/10.1038/nature04643
Lin-Chao S, Chen WT, Wong TT (1992) High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol Microbiol 6:3385–3393. https://doi.org/10.1111/j.1365-2958.1992.tb02206.x
Chang AC, Cohen SN (1978) Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant No. 81572034, the Development Fund for Shanghai Talents under Grant No. 2018098, Fengxian District (Society) Science and Technology Development Fund Project under Grant No. 20151001.
Author information
Authors and Affiliations
Contributions
QW and XW conceived the study. GL coordinated the study. XW, NK, MC, LZ and MS performed the experiments. XW and QW analyzed the data and wrote the manuscript. QW and GL revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Neither author has any conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Kong, N., Cao, M. et al. Comparison of Class 2 Integron Integrase Activities. Curr Microbiol 78, 967–978 (2021). https://doi.org/10.1007/s00284-021-02352-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02352-9