Abstract
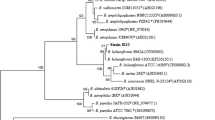
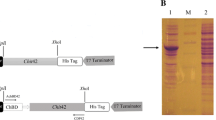
The chitinases are gaining much attention based on their role in the defense against pathogen attacks and harmful insects. The partially chitinase produced by Bacillus licheniformis strain J24 exhibited a large antifungal spectrum, and the highest activity was obtained toward Fusarium species in vitro on PDA and in vivo on corn seeds. The chitinase was inducible by the presence of autoclaved Fusarium conidia in the medium culture and it was active at 70 °C and pH 7 and not affected by the tested chemical agents EDTA and SDS. The nucleotide and amino acid sequences encoding chitinase showed the close phylogenetic relation with chitinase from Bacillus paralicheniformis species. Based on the analysis of the putative domain active, the described chitinase from strain J24 was belonging to the GH family-18 and the novelty of its structure was revealed. Here the combination of functional and structural antifungal extremely chitinase proves its importance in biotechnology area.





Similar content being viewed by others
References
Liu D, Caia J, Xie C, Liu C, Chen Y (2010) Purification and partial characterization of a 36 kDa chitinase from Bacillus thuringiensis subsp colmeri and its biocontrol potential. Enzyme Microbial Technol 46:252–256
Swiontek BM, Jankiewicz U, Burkowska A, Walczak M (2014) Chitinolytic microorganisms ans their possible application in environmental protection. Curr Microbiol 68:71–81
Nampoothiri MK, Baiju TV, Sandhya C, Sabu A, Szakacs G et al (2004) Process optimization for antifungal chitinase production by Trichoderma harzianum. Process Biochem 39:1583–1590
Matroudi S, Zamani MR, Motallebi M (2008) Molecualr cloning of chitinase 33 (chit 33) gene from Trichoderma atroviride. Braz J Microbiol 39:433–437
Essghaier B, Hedi A, Bejji M, Jijakli H, Boudabous et al (2011) Characterization of a novel chitinase from a moderately halophilic bacterium Virgibacillus marismortui strain M3-23. Ann Microbiol 62(2): 835–841
Ghorbel-Bellaaj O, Manni L, Jellouli K, Hmidet N, Nasri M (2012) Optimization of protease and chitinase production by Bacillus cereus SV1 on shrimp shell waste using statistical experimental design. Biochemical and molecular characterization of the chitinase. Ann Microbiol 62:1255–1268
Waldeck J, Daum G, Bisping B, Meinhardt F (2006) Isolation and molecular characterization of chitinase deficient Bacillus licheniformis strains capable of deproteinization of shrimp shell waste to obtain highly viscous chitin. Appl Environ Microbiol 72:7879–7885
Ohnuma T, Sorlie M, Fukuda T, Kawamato N, Taira T, Fukamizo T (2011) Chitin oligosaccharide binding to a family GH19 chitinase from the moss Bryum coronatum. FEBS J 278:3991–4001
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293(3):781–788
Duan C, Qin Z, Yang Z, Li W, Sun S et al (2016) Identification of pathogenic Fusarium spp causing maize ear rot and potential mycotoxin production in China. Toxins 8(6):186
McGovern RJ (2015) Management of tomato diseases caused by Fusarium oxysporum. Crop Protect 73:78–92
Khan N, Maymon M, Hirsch AN (2017) Combating fusarium infection using bacillus-based antimicrobials. Microorganisms 5:75
Essghaier B, Fardeau ML, Cayol JL, Hajlaoui MR, Boudabous A et al (2009a) Biological control of grey mold in strawberry fruits by halophilic bacteria. J App Microbiol 106(3):833–846
Gomez Ramirez M, Rojas Avelizapa LI, Rojas Avelizapa NG, Cruz Camarillo R (2004) Colloidal chitin stained with Remazol Brillant blue R, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J Microbiol Methods 56(2):213–219
Issam SM, Mohamed G, Farid L, Sami F, Thierry M, et al (2003) Production, purification and biochemical characterization of two beta-glucosidases from Sclerotinia sclerotiorum. Appl Biochem Biotechnol 111 (1): 29–40
Ellouze O, Mejri M, Smaali I, Limam F, Marzouki MN (2007) Induction, properties and application of xylmanase activity from Sclerotinia sclerotiorum S2 Fungus. J Food Biochem 31:96–107
Essghaier B, Bejji M, Jijakli H, Boudabous A, Sadfi-Zouaoui N (2009b) High salt-tolerant protease from a potential biocontrol agent Bacillus pumilus M3-16. Ann Microbiol 59(3):553–558
Altschul SF, Madden TL, Schaffer AA, Zhang Z, Miller W et al (1997) Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Taylor G, Jabaji-hare S, Charest PM, Khan W (2002) Purification and characterization of an extracellular exochitinase, β-N-acdetylhexosaminidase, from the fungal mycoparasitte Stachybotrys elegans. Can J Microbiol 48:311–311
Thamthiankul S, Suan-Ngay S, Tantimavanich S, Panbangred W (2001) Chitinase from Bacillus thuringiensis subsp. Pakistani. Appl Microbiol Biotechnol 56:396–401
Swiontek BM, Jankiewicz U, Lisiecki K (2013) Optimization of cultural conditions for the production of antifungal chitinase by Streptomyces sporovirgulis. Appl Biochem Microbiol 49(2):154–159
Chang WT, Chen YC, Jao CL (2007) Antifungal activity and enhancement of plant growth by Bacillus cereus grown on shellfish chitin wastes. Bioresour Technol 98:1224–1230
Ghasemi S, Ahmadian G, Jelodar NB, Rahimian H, Ghandili S et al (2010) Antifungal chitinases from Bacillus pumilus SG-2: preliminary report. World J Microbiol Biotechnol 26:1437–1443
Dahiya N, Tewari RP, Hoondal GS (2005) Production of an antifungal chitianse from Enterobacter sp NRG4 and its application in protoplast production. World J Microbiol Biotechnol 21:1611–1616
Jankiewicz U, Swiontek BM, Saks E (2012) Identification and characterization of a chitinase of Stenotrophomonas maltophilia, a bacterium that is antagonistic towards fungal phytopathogens. J Biosci Bioeng 113(1):30–35
Khiyami M, Masmali I (2008) Characteristics of thermostable chitinase enzymes of Bacillus licheniformis isolated from Red Palm Weavil Gut. Aus J Basic Appl Sci 2(4):943–948
Yuli PE, Suhartono MT, Rukayadi Y, Hwang JK, Pyun YR (2004) Characteristics of thermostable chitinase enzymes from the Indonesian Bacillus sp 13.26. Enzyme Microbial Technol 35:147–153
Kudan S, Rath P (2009) Purification and characterization of termostable chitinase from Bacillus licheniformis SK-1. Appl Biochem Biotechnol 157:23–35
Lombard V, Ramulu HG, Drula E, Corytinho PM, Henrissat B (2014) The carbohydrate active enzymes CAZY in 2013. Nucleic Acids Res 42:490–495 (http://www.cazy.org/)
Arguelles Arias A, Ongena M, Halimi B, Lara Y, Brans A et al (2009) Bacillus amyloquefasciens GA 1as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb Cell Factories 8(63)
Essghaier B, Rouaissi M, Boudabous A, Jijakli H, Sadfi-Zouaoui N (2010) Production and partial characterization of chitinase from a halotolerant Planococcus rifitoensis strain M2-26. World J Microbiol Biotechnol 26:977–984
Acknowledgements
This work was supported by funds from the Ministry of Higher Education and Scientific Research of Tunisia (LR16ES05).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Essghaier, B., Zouaoui, M., Najjari, A. et al. Potentialities and Characterization of an Antifungal Chitinase Produced by a Halotolerant Bacillus licheniformis. Curr Microbiol 78, 513–521 (2021). https://doi.org/10.1007/s00284-020-02329-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02329-0