Abstract
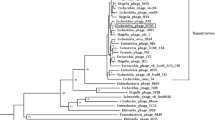
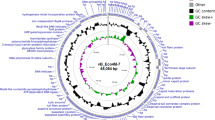
Escherichia coli O157:H7 is an important foodborne pathogen that has become a major worldwide factor affecting the public safety of food. Bacteriophage has gradually attracted attention because of its ability to kill specific pathogens. In this study, a lytic phage of E. coli O157:H7, named FEC14, was isolated from hospital sewage. Transmission electron microscopy analysis showed that phage FEC14 had an isometric head 80 ± 5 nm in diameter and a contractile tail whose terminal spikes present an umbrella-like structure. Phage FEC14 revealed 158,639 bp double-stranded DNA, with the G+C content of 44.6%, 209 ORFs and four tRNAs. Genome DNA of FEC14 could not be digested by some endonucleases. Many of the features of phage FEC14 are very similar to those of the newly classified genus “Kuttervirus”, including morphology, genome size and organization, etc. Phage FEC14 is proposed to be a new isolate of genus “Kuttervirus” within the family Ackermannviridae, moreover, the endonuclease resistance of phage FEC14, has priority over other genera of bacteriophages for its use in biocontrol of foodborne pathogens.






Similar content being viewed by others
References
Carroll KC, Morse ST, Mietzner T, Miller S (2016) Jawetz, Melnick & Adelberg’s Medical Microbiology, 27th edn. McGraw-Hill Education, New York
Chapman PA, Cerdan Malo AT, Ellin M, Ashton R, Harkin MA (2001) Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. Int J of Food Microbiol 119:245–250
Saxena T, Kanshik P, Mohan MK (2015) Prevalence of E. Coli O157:H7 in water sources: an overview on associated diseases, outbreaks and detection methods. Diagn Micr Infect Dis 82:249–264
Chapman PA, Siddons CA, Harkin MA (1996) Sheep as a potential source of verocytotoxin-producing Escherichia coli O157. Vet Rec 138:23–24
Carter CD, Parks A, Abuladze T, Li M, Woolston J et al (2012) Bacteriophage cocktail significantly reduces Escherichia coli O157: H7 contamination of lettuce and beef, but does not protect against recontamination. Bacteriophage 2:178–185
Coffey B, Rivas L, Duffy G, Coffey A, Ross RP et al (2011) Assessment of Escherichia coli O157:H7-specific bacteriophages e11/2 and e4/1c in model broth and hide environments. Int J Food Microbiol 147:188–194
Bigwood T, Hudson JA, Billington C (2009) Influence of host and bacteriophage concentrations on the inactivation of food-borne pathogenic bacteria by two phages. FEMS Microbiol Lett 291:59–64
Guenther S, Huwyler D, Richard S, Loessner MJ (2009) Virulent bacteriophage for efficient biocontrol of Lister monocytogenes in ready-to-eat foods. Appl Environ Microbiol 75:93–100
Moye ZD, Woolston J, Sulakvelize A (2018) Bacteriophage applications for food production and processing. Virus 10:205
Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP (2009) Enumeration of bacteriophages by double agar overlay plaque assay. In: Clokie M (ed) Bacteriophages. Humana Press, New Jersey, pp 69–76
Fan N, Qi R, Yang M (2017) Isolation and characterization of a virulent bacteriophage infecting Acinetobacter johnsonii from activated sludge. Res Microbiol 168:472–481
Kutter E (2009) Phage host range and efficiency of plating. In: Clokie M (ed) Bacteriophages. Humana Press, New Jersey, pp 141–149
Nasukawa T, Uchiyama J, Taharaguchi S, Ota S, Ujihara T et al (2017) Virus purification by CsCl density gradient using general centrifugation. Arch Virol 162:3523–3528
Petrovski S, Seviour RJ, Tillett D (2011) Genome sequence and characterization of the Tsukamurella bacteriophage TPA2. Appl Environ Microbiol 77:1389–1398
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54-57
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and mRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Chen L, Yang J, Yu J, Yao Z, Sun L et al (2005) VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P et al (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010
Kropinski AM, Bose RJ, Warren RA (1973) 5-(4-Aminobutylaminomethyl) uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage phiW-14. Biochemistry 12:151–157
Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL et al (2009) The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 388:48–70
Hoet PP, Coene MM, Cocito CG (1992) Replication cycle of Bacillus subtilis hydroxymethyluracil-containing phages. Annu Rev Microbiol 46:95–116
Plattner M, Shneider MM, Arbatsky NP, Shashkov AS, Chizhov AO et al (2019) Structure and function of the branched receptor-binding complex of bacteriophage CBA120. J Mol Biol 431:3718–3739
Park M, Lee JH, Shin H, Kim M, Choi J et al (2012) Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol 78:58–69
Hooton SP, Timms AR, Rowsell J, Wilson R, Connerton IF (2011) Salmonella Typhimurium-specific bacteriophage PhiSH19 and the origins of species specificity in the Vi01-like phage family. Virol J 8:498
Karpe YA, Kanade GD, Pingale KD, Arankalle VA, Banerjee K (2016) Genomic characterization of Salmonella bacteriophages isolated from India. Virus Genes 52:117–126
Adriaenssens EM, Ackermann HW, Anany H, Blasdel B, Connerton IF et al (2012) A suggested new bacteriophage genus: “Viunalikevirus.” Arch Virol 157:2035–2046
Wang Y, Wang W, Lv Y, Zheng W, Mi Z et al (2014) Characterization and complete genome sequence analysis of novel bacteriophage IME-EFm1 infecting Enterococcus faecium. J General Virol 95:2565–2575
Gong Z, Wang M, Yang Q, Li Z, Xia J et al (2017) Isolation and complete genome sequence of a novel Pseudoalteromanoa phage PH357 from the Yangtze River estuary. Curr Microbiol 74:832–839
Chen C, Bales P, Greenfield J, Heselpoth RD, Nelson DC et al (2014) Crystal structure of ORF210 from E. coli O157:H7 phage CBA120 (TSP1), a putative tailspike protein. PLoS ONE 9:e93156
Kutter EM, Skutt-Kakaria K, Blasdel B, El-Shibiny A, Castano A et al (2011) Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J 8:430
Maltman KL, Neuhard J, Warren RA (1981) 5-[(Hydroxymethyl)-O-pyrophosphoryl] uracil, an intermediate in the biosynthesis of alpha-putrescinylthymine in deoxyribonucleic acid of bacteriophage phi W-14. Biochemistry 20:3586–3591
Pleska M, Qian L, Okura R, Bergmiller T, Wakamoto Y et al (2016) Bacterial autoimmunity due to a restriction-modification system. Curr Biol 26:404–409
Acknowledgements
The authors thank Dr. Yanbo Sun (Department of Pathogenobiology, College of Basic Medical Sciences, Jilin University) for constructive comments. This reseach was supported by a grant from Health Comission of Jilin Province (No. 2020J032).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fan, C., Tie, D., Sun, Y. et al. Characterization and Genomic Analysis of Escherichia coli O157:H7 Bacteriophage FEC14, a New Member of Genus Kuttervirus. Curr Microbiol 78, 159–166 (2021). https://doi.org/10.1007/s00284-020-02283-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02283-x