Abstract
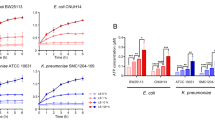
Persisters of infectious agents are capable of surviving antibiotic treatment so the emergence of these subpopulations need to be overcome. In this study, we aimed to isolate, characterize and inhibit persister subpopulation in two clinical isolates Klebsiella pneumoniae and Proteus mirabilis. Different behavior profiles between the two isolates could be observed. The results of dose-dependent killing curve revealed that 2.3% (Klebsiella pneumoniae) versus 1.3% (Proteus mirabilis) persister cells could be recovered using 500 and 30 ug/ml ciprofloxacin, respectively. Upon resuscitation, persister cells exhibited only 65% versus 30% percentage growth and 5 versus 7 times cell elongation relative to Klebsiella pneumoniae and Proteus mirabilis, respectively. The levels of persister cells to ciprofloxacin of Klebsiella pneumoniae were dramatically decreased by about 79, 92, 97 and 83% in average by pre-exposure to hyperosmotic stress, temperature, different pHs, and hydrogen peroxide, respectively, while those of Proteus mirabilis were minimally decreased with corresponding reduction percentages of about 12%, 24 & 25%, and 0%. Regarding combating persisters, Klebsiella pneumoniae showed different response as compared to Proteus mirabilis. Among the tested sugars, the highest reduction of Klebsiella pneumoniae persister cells was obtained with pre-priming with sucrose while for Proteus mirabilis persister cells, the highest reduction was obtained with pre-priming with glucose. Using sodium salicylate with ciprofloxacin could eradicate persisters of Klebsiella pneumoniae at any tested concentration while for Proteus mirabilis it caused some reduction in persister cells at certain concentrations. Complete eradication of persisters was obtained by combining silver nitrate with ciprofloxacin for each test isolate.
Similar content being viewed by others
References
Zhang Y (2014) Persisters, persistent infections and the Yin-Yang model. Emerg Microbes Infect 3:e3. https://doi.org/10.1038/emi.2014.3
Allison KR, Brynildsen MP, Collins JJ (2011) Heterogeneous bacterial persisters and engineering approaches to eliminate them. Curr Opin Microbiol 14:593–598. https://doi.org/10.1016/j.mib.2011.09.002
Cohen NR, Lobritz MA, Collins JJ (2013) Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. https://doi.org/10.1016/j.chom.2013.05.009
Bigger JW (1944) The bactericidal action of penicillin on staphylococcus pyogenes. Ir J Med Sci 19:585–595. https://doi.org/10.1007/BF02948462
Spoering AL, Lewis K (2001) Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. https://doi.org/10.1128/JB.183.23.6746-6751.2001
Lewis K (2005) Persister cells and the riddle of biofilm survival. Biochemistry (Moscow) 70:267–274. https://doi.org/10.1007/s10541-005-0111-6
Dörr T, Lewis K, Vulić M (2009) SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genetics 5:e1000760. https://doi.org/10.1371/journal.pgen.1000760
Wu Y, Vulić M, Keren I, Lewis K (2012) Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. https://doi.org/10.1128/AAC.00921-12
Hong SH, Wang X, O’Connor HF et al (2012) Bacterial persistence increases as environmental fitness decreases. Microb Biotechnol 5:509–522. https://doi.org/10.1111/j.1751-7915.2011.00327.x
Kwan BW, Valenta JA, Benedik MJ, Wood TK (2013) Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473
Lewis K (2012) Persister cells: molecular mechanisms related to antibiotic tolerance. Antibiotic Resistance. Springer, Berlin, Heidelberg, pp 121–133
Kwan BW, Chowdhury N, Wood TK (2015) Combatting bacterial infections by killing persister cells with mitomycin C. Environ Microbiol 17:4406–4414. https://doi.org/10.1111/1462-2920.12873
CLSI (2014) Performance Standards for Antimicrobials Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S25 Wayne, PA: Clinical and Laboratory Standards Institute
Niepa THR, Gilbert JL, Ren D (2012) Controlling Pseudomonas aeruginosa persister cells by weak electrochemical currents and synergistic effects with tobramycin. Biomaterials 33:7356–7365
Pascoe B, Dams L, Wilkinson S et al (2014) Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS ONE 9:e85998. https://doi.org/10.1371/journal.pone.0085998
Grassi L, Di Luca M, Maisetta G et al (2017) Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front Microbiol. https://doi.org/10.3389/fmicb.2017.01917
Abokhalil RN, Elkhatib WF, Aboulwafa MM, Hassouna NA (2018) Recovery and characterization of Proteus mirabilis persisters. Archiv Pharm Sci Ain Shams Univ 2:31–36. https://doi.org/10.21608/aps.2018.18732
Fischer ER, Hansen BT, Nair V et al (2012) Scanning electron microscopy. Curr Protoc Microbiol Chap. https://doi.org/10.1002/9780471729259.mc02b02s25
Ma C, Sim S, Shi W et al (2010) Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol Lett 303:33–40. https://doi.org/10.1111/j.1574-6968.2009.01857.x
Allison KR, Brynildsen MP, Collins JJ (2011) Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. https://doi.org/10.1038/nature10069
Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ (2013) Sci Transl Med. https://doi.org/10.1126/scitranslmed.3006276
Wang T, El Meouche I, Dunlop MJ (2017) Bacterial persistence induced by salicylate via reactive oxygen species. Sci Rep. https://doi.org/10.1038/srep43839
Li Y, Zhang L, Zhou Y et al (2018) Survival of bactericidal antibiotic treatment by tolerant persister cells of Klebsiella pneumoniae. J Med Microbiol 67:273–281. https://doi.org/10.1099/jmm.0.000680
Bhardwaj R, Qamar I, Singh N (2015) Persister formation in Klebsiella pneumoniae: recombinant expression and Purification of hipA and hipB proteins from K. pneumoniae. International journal of applied and pure science and agriculture 1:1–7
Boonsarngsuk V, Thungtitigul P, Suwatanapongched T (2015) Chronic Klebsiella pneumonia: a rare manifestation of Klebsiella pneumonia. J Thorac Dis 7:1661–1664. https://doi.org/10.3978/j.issn.2072-1439.2015.09.31
Hobby GL, Meyer K, Chaffee E (2016) Observations on the mechanism of action of penicillin. Soc Exp Biol Med 50:281–285. https://doi.org/10.3181/00379727-50-13773
Möker N, Dean C, Tao J (2010) Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J Bacteriol 192:1946–2195
Goneau LW, Yeoh NS, MacDonald KW et al (2014) Selective target inactivation rather than global metabolic dormancy causes antibiotic tolerance in uropathogens. Antimicrob Agents Chemother 58:2089–2097. https://doi.org/10.1128/AAC.02552-13
Ren H, He X, Zou X et al (2015) Gradual increase in antibiotic concentration affects persistence of Klebsiella pneumoniae. J Antimicrob Chemother 70:3267–3272. https://doi.org/10.1093/jac/dkv251
Keren I, Kaldalu N, Spoering A et al (2004) Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18
Maisonneuve E, Castro-Camargo M, Gerdes K (2013) (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150
Prax M, Bertram R (2014) Metabolic aspects of bacterial persisters. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2014.00148
Kaldalu N, Hauryliuk V, Tenson T (2016) Persisters—as elusive as ever. Appl Microbiol Biotechnol 100:6545–6553. https://doi.org/10.1007/s00253-016-7648-8
Renbarger TL, Baker JM, Sattley WM (2017) Slow and steady wins the race: an examination of bacterial persistence. Microbiology 3:171–185. https://doi.org/10.3934/microbiol.2017.2.171
Wood TK, Knabel SJ, Kwan BW (2013) Bacterial Persister Cell Formation and Dormancy. Appl Environ Microbiol 79:7116–7121. https://doi.org/10.1128/AEM.02636-13
Kim J-S, Yamasaki R, Song S, et al Single cell observations show persister cells wake based on ribosome content. Environ Microb. https://doi.org/10.1111/1462-2920.14093
Balaban NQ, Merrin J, Chait R et al (2004) Bacterial persistence as a phenotypic switch. Science 305:1622–1625. https://doi.org/10.1126/science.1099390
Cho J, Carr AN, Whitworth L et al (2017) MazEF toxin-antitoxin proteins alter Escherichia coli cell morphology and infrastructure during persister formation and regrowth. Microbiology 163:308–321. https://doi.org/10.1099/mic.0.000436
Li B, Qiu Y, Glidle A et al (2014) Single cell growth rate and morphological dynamics revealing an “opportunistic” persistence. Analyst 139:3305–3313. https://doi.org/10.1039/C4AN00170B
Martinez OV, Gratzner HG, Malinin TI, Ingram M (1982) The effect of some beta-lactam antibiotics on Escherichia coli studied by flow cytometry. Cytometry 3:129–133. https://doi.org/10.1002/cyto.990030211
Chung HS, Yao Z, Goehring NW et al (2009) Rapid β-lactam-induced lysis requires successful assembly of the cell division machinery. PNAS 106:21872–21877. https://doi.org/10.1073/pnas.0911674106
Yao Z, Kahne D, Kishony R (2012) Distinct single-cell morphological dynamics under beta-lactam antibiotics. Molecular Cell 48:705–712. https://doi.org/10.1016/j.molcel.2012.09.016
Fisher RA, Gollan B, Helaine S (2017) Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. https://doi.org/10.1038/nrmicro.2017.42
Leung V, Lévesque CM (2012) A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol 194:2265–2274. https://doi.org/10.1128/JB.06707-11
Lewis K (2007) Persister cells, dormancy and infectious disease. Nat Rev Microb 5:48–56. https://doi.org/10.1038/nrmicro1557
Wood KB, Cluzel P (2012) Trade-offs between drug toxicity and benefit in the multi-antibiotic resistance system underlie optimal growth of E. coli. BMC Syst Biol 6:48. https://doi.org/10.1186/1752-0509-6-48
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or nonprofit sectors.
Author information
Authors and Affiliations
Contributions
RA: Design and conduction of experiments, data collection and presentation, draft writing of the manuscript. WE: Experimental design, data analysis, interpretation, supervision, revise manuscript. MA: Conception of the research idea, supervision, interpretation of data, manuscript writing and final editing. NH: Data analysis and interpretation, supervision, manuscript revision. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abokhalil, R.N., Elkhatib, W.F., Aboulwafa, M.M. et al. Persisters of Klebsiella pneumoniae and Proteus mirabilis: A Common Phenomenon and Different Behavior Profiles. Curr Microbiol 77, 1233–1244 (2020). https://doi.org/10.1007/s00284-020-01926-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01926-3