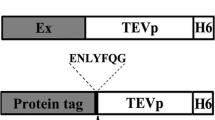
Abstract
Expression and secretion of recombinant proteins in the endotoxin-free bacterium, Bacillus subtilis, has been thoroughly studied, but overexpression in the cytoplasm has been limited to only a few proteins. Here, we used the robust IPTG-inducible promoter, Pgrac212, to overexpress human rhinovirus 3C protease (HRV3C) in the cytoplasm of B. subtilis cells. A novel solubility tag, the N-terminal domain of the lysS gene of B. subtilis coding for a lysyl-tRNA synthetase was placed at the N terminus with a cleavage site for the endoprotease HRV3C, followed by His-HRV3C or His-GST-HRV3C. The recombinant protease was purified by using a Ni–NTA column. In this study, the His-HRV3C and His-GST-HRV3C proteases were overexpressed in the cytoplasm of B. subtilis at 11% and 16% of the total cellular proteins, respectively. The specific protease activities were 8065 U/mg for His-HRV3C and 3623 U/mg for His-GST-HRV3C. The purified enzymes were used to cleave two different substrates followed by purification of the two different protein targets, the green fluorescent protein and the beta-galactosidase. In conclusion, the combination of an inducible promoter Pgrac212 and a solubility tag allowed the overexpression of the HRV3C protease in the cytoplasm of B. subtilis. The resulting fusion protein was purified using a nickel column and was active in cleaving target proteins to remove the fusion tags. This study offers an effective method for producing recombinant proteins in the cytoplasm of endotoxin-free bacteria.




Similar content being viewed by others
References
Westers L, Westers H, Quax WJ (2004) Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim Biophys Acta 1694:299–310. https://doi.org/10.1016/j.bbamcr.2004.02.011
Zweers JC, Barák I, Becher D et al (2008) Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb Cell Fact 7:10. https://doi.org/10.1186/1475-2859-7-10
Harwood CR, Cranenburgh R (2008) Bacillus protein secretion: an unfolding story. Trends Microbiol 16:73–79. https://doi.org/10.1016/j.tim.2007.12.001
Pohl S, Harwood CR (2010) Heterologous protein secretion by Bacillus species from the cradle to the grave. Adv Appl Microbiol 73:1–25. https://doi.org/10.1016/S0065-2164(10)73001-X
Wong SL (1995) Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr Opin Biotechnol 6:517–522
Shields DC, Sharp PM (1987) Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res 15:8023–8040
Kang Z, Yang S, Du G, Chen J (2014) Molecular engineering of secretory machinery components for high-level secretion of proteins in Bacillus species. J Ind Microbiol Biotechnol 41:1599–1607. https://doi.org/10.1007/s10295-014-1506-4
Schallmey M, Singh A, Ward OP (2004) Developments in the use of Bacillus species for industrial production. Can J Microbiol 50:1–17. https://doi.org/10.1139/w03-076
Schumann W (2007) Production of recombinant proteins in Bacillus subtilis. Adv Appl Microbiol 62:137–189. https://doi.org/10.1016/S0065-2164(07)62006-1
van Dijl JM, Hecker M (2013) Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb Cell Fact 12:3. https://doi.org/10.1186/1475-2859-12-3
Heinrich J, Drewniok C, Neugebauer E et al (2019) The YoaW signal peptide directs efficient secretion of different heterologous proteins fused to a StrepII-SUMO tag in Bacillus subtilis. Microb Cell Fact 18:31. https://doi.org/10.1186/s12934-019-1078-0
Taguchi S, Ooi T, Mizuno K, Matsusaki H (2015) Advances and needs for endotoxin-free production strains. Appl Microbiol Biotechnol 99:9349–9360. https://doi.org/10.1007/s00253-015-6947-9
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. https://doi.org/10.3389/fmicb.2014.00172
Zhao Y, He W, Liu W-F et al (2012) Two distinct states of Escherichia coli cells that overexpress recombinant heterogeneous β-galactosidase. J Biol Chem 287:9259–9268. https://doi.org/10.1074/jbc.M111.327668
Cui W, Han L, Cheng J et al (2016) Engineering an inducible gene expression system for Bacillus subtilis from a strong constitutive promoter and a theophylline-activated synthetic riboswitch. Microb Cell Fact 15:199. https://doi.org/10.1186/s12934-016-0599-z
Nguyen NH, Phan TTP, Tran TL, Nguyen HD (2014) Investigating the expression of GFP fused with HIS-TAG at N- or C-terminus using plasmid pHT253 and pHT254 in Bacillus subtilis. Sci Technol Dev J 17(4), 5–11. 10.32508/stdj.v17i4.1550.
Phan TTP, Tran LT, Schumann W, Nguyen HD (2015) Development of Pgrac100-based expression vectors allowing high protein production levels in Bacillus subtilis and relatively low basal expression in Escherichia coli. Microb Cell Fact 14:72. https://doi.org/10.1186/s12934-015-0255-z
Wenzel M, Müller A, Siemann-Herzberg M, Altenbuchner J (2011) Self-inducible Bacillus subtilis expression system for reliable and inexpensive protein production by high-cell-density fermentation. Appl Environ Microbiol 77:6419–6425. https://doi.org/10.1128/AEM.05219-11
Phan TTP, Nguyen HD, Schumann W (2012) Development of a strong intracellular expression system for Bacillus subtilis by optimizing promoter elements. J Biotechnol 157:167–172. https://doi.org/10.1016/j.jbiotec.2011.10.006
Phan T, Huynh P, Truong T, Nguyen H (2017) A generic protocol for intracellular expression of recombinant proteins in Bacillus subtilis. Methods Mol Biol 1586:325–334. https://doi.org/10.1007/978-1-4939-6887-9_21
Tran DTM, Phan TTP, Huynh TK et al (2017) Development of inducer-free expression plasmids based on IPTG-inducible promoters for Bacillus subtilis. Microb Cell Fact 16:130. https://doi.org/10.1186/s12934-017-0747-0
Phan TTP, Nguyen HD, Schumann W (2006) Novel plasmid-based expression vectors for intra- and extracellular production of recombinant proteins in Bacillus subtilis. Protein Expr Purif 46:189–195. https://doi.org/10.1016/j.pep.2005.07.005
Phan TTP, Nguyen ALT, Nguyen HD (2013) Cloning and expression of LTB in Escherichia coli and Bacillus subtilis. Nat Sci 16(1), 13–22. https://doi.org/10.32508/stdj.v16i1.1392
Ngo HK, Nguyen ALT, Huynh PTK, et al. (2016) Expression and purification of listeriolysin O from Listeria monocytogenes harbouring E247M and D320K mutations in Bacillus subtilis. Sci Technol Dev J 19:20–31. https://doi.org/10.32508/stdj.v19i3.470
Truong TTT, Phan TTP, Nguyen HD (2017) Purification of P24 protein expressed in Bacillus subtilis and evaluation of its immunogenicity in mice. Sci Technol Dev J 1:69–79. https://doi.org/10.32508/stdjns.v1iT1.436
Zukowski MM, Miller L (1986) Hyperproduction of an intracellular heterologous protein in a sacUh mutant of Bacillus subtilis. Gene 46:247–255
Peschke U, Beuck V, Bujard H et al (1985) Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J Mol Biol 186:547–555
Puohiniemi R, Butcher S, Tarkka E, Sarvas M (1991) High level production of Escherichia coli outer membrane proteins OmpA and OmpF intracellularly in Bacillus subtilis. FEMS Microbiol Lett 67:29–33. https://doi.org/10.1111/j.1574-6968.1991.tb04383.x
Titok MA, Chapuis J, Selezneva YV et al (2003) Bacillus subtilis soil isolates: plasmid replicon analysis and construction of a new theta-replicating vector. Plasmid 49:53–62
Nguyen HD, Nguyen QA, Ferreira RC et al (2005) Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid 54:241–248. https://doi.org/10.1016/j.plasmid.2005.05.001
Nguyen HD, Phan TTP, Schumann W (2007) Expression vectors for the rapid purification of recombinant proteins in Bacillus subtilis. Curr Microbiol 55:89–93. https://doi.org/10.1007/s00284-006-0419-5
Phan TTP, Nguyen HD, Schumann W (2010) Establishment of a simple and rapid method to screen for strong promoters in Bacillus subtilis. Protein Expr Purif 71:174–178. https://doi.org/10.1016/j.pep.2009.11.010
Phan TTP, Nguyen HD, Schumann W (2013) Construction of a 5′-controllable stabilizing element (CoSE) for over-production of heterologous proteins at high levels in Bacillus subtilis. J Biotechnol 168:32–39. https://doi.org/10.1016/j.jbiotec.2013.07.031
Trang PTP (2007) Construction and analysis of novel controllable expression vectors for Bacillus subtilis. Doctoral thesis, University of Bayreuth, Bayreuth
Young CL, Britton ZT, Robinson AS (2012) Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol J 7:620–634. https://doi.org/10.1002/biot.201100155
Waugh DS (2011) An overview of enzymatic reagents for the removal of affinity tags. Protein Expr Purif 80:283–293. https://doi.org/10.1016/j.pep.2011.08.005
Walker PA, Leong LE, Ng PW et al (1994) Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Biotechnology (NY) 12:601–605
Vergis JM, Wiener MC (2011) The variable detergent sensitivity of proteases that are utilized for recombinant protein affinity tag removal. Protein Expr Purif 78:139–142. https://doi.org/10.1016/j.pep.2011.04.011
Antoniou G, Papakyriacou I, Papaneophytou C (2017) Optimization of soluble expression and purification of recombinant human rhinovirus type-14 3C protease using statistically designed experiments: isolation and characterization of the enzyme. Mol Biotechnol 59:407–424. https://doi.org/10.1007/s12033-017-0032-9
Cordingley MG, Callahan PL, Sardana VV et al (1990) Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J Biol Chem 265:9062–9065
Raran-Kurussi S, Tözsér J, Cherry S et al (2013) Differential temperature dependence of tobacco etch virus and rhinovirus 3C proteases. Anal Biochem 436:142–144. https://doi.org/10.1016/j.ab.2013.01.031
Choi SI, Han KS, Kim CW et al (2008) Protein solubility and folding enhancement by interaction with RNA. PLoS ONE 3:e2677. https://doi.org/10.1371/journal.pone.0002677
Horowitz S, Bardwell JCA (2016) RNAs as chaperones. RNA Biol 13:1228–1231. https://doi.org/10.1080/15476286.2016.1247147
Yang SW, Jang YH, Kwon SB et al (2018) Harnessing an RNA-mediated chaperone for the assembly of influenza hemagglutinin in an immunologically relevant conformation. FASEB J 32:2658–2675. https://doi.org/10.1096/fj.201700747RR
Feng Y, Xu Q, Yang T et al (2014) A novel self-cleavage system for production of soluble recombinant protein in Escherichia coli. Protein Expr Purif 99:64–69. https://doi.org/10.1016/j.pep.2014.04.001
Xu H, Wang Q, Zhang Z et al (2019) A simplified method to remove fusion tags from a xylanase of Bacillus sp. HBP8 with HRV 3C protease. Enzyme Microb Technol 123:15–20. https://doi.org/10.1016/j.enzmictec.2019.01.004
Paccez JD, Nguyen HD, Luiz WB et al (2007) Evaluation of different promoter sequences and antigen sorting signals on the immunogenicity of Bacillus subtilis vaccine vehicles. Vaccine 25:4671–4680. https://doi.org/10.1016/j.vaccine.2007.04.021
Cui W, Han L, Suo F et al (2018) Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J Microbiol Biotechnol 34:145. https://doi.org/10.1007/s11274-018-2531-7
Öztürk S, Ergün BG, Çalık P (2017) Double promoter expression systems for recombinant protein production by industrial microorganisms. Appl Microbiol Biotechnol 101:7459–7475. https://doi.org/10.1007/s00253-017-8487-y
Song Y, Nikoloff JM, Zhang D (2015) Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis. J Microbiol Biotechnol 25:963–977. https://doi.org/10.4014/jmb.1501.01028
Westers H, Dorenbos R, van Dijl JM et al (2003) Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol Biol Evol 20:2076–2090. https://doi.org/10.1093/molbev/msg219
Ara K, Ozaki K, Nakamura K et al (2007) Bacillus minimum genome factory: effective utilization of microbial genome information. Biotechnol Appl Biochem 46:169–178. https://doi.org/10.1042/BA20060111
Aguilar Suárez R, Stülke J, van Dijl JM (2019) Less is more: toward a genome-reduced Bacillus cell factory for “difficult proteins”. ACS Synth Biol 8:99–108. https://doi.org/10.1021/acssynbio.8b00342
Saito H, Shibata T, Ando T (1979) Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol Gen Genet 170:117–122
Funding
This research was funded by the Department of Science and Technology of Ho Chi Minh City, Vietnam under Grant Number 1022/QĐ-SKHCN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Research Involving Human and Animal Rights
This study did not use animals or samples from human for experiments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Le, V.D., Phan, T.T.P., Nguyen, T.M. et al. Using the IPTG-Inducible Pgrac212 Promoter for Overexpression of Human Rhinovirus 3C Protease Fusions in the Cytoplasm of Bacillus subtilis Cells. Curr Microbiol 76, 1477–1486 (2019). https://doi.org/10.1007/s00284-019-01783-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01783-9