Abstract
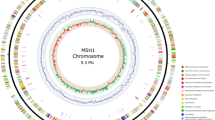
Carbaryl was a widely used pesticide in the agriculture industry. The toxicity against non-target organisms and the environmental pollution it caused became the focus of public concern. However, the microbial mechanism of carbaryl degradation was not fully investigated. In the study, we reported the complete genome of the carbaryl-degrading Pseudomonas putida strain XWY-1, which consists of a chromosome (5.9 Mbp) and a plasmid (0.4 Mbp). The carbaryl degradation genes are located on the plasmid. The study on the genome will facilitate to further elucidate the carbaryl degradation and advance the potential biotechnological applications of P. putida strain XWY-1.


Similar content being viewed by others
References
Zylstra G, McCombie W, Gibson D, Finette B (1988) Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol 54(6):1498–1503
Abuhamed T, Bayraktar E, Mehmetoğlu T, Mehmetoğlu Ü (2004) Kinetics model for growth of Pseudomonas putida F1 during benzene, toluene and phenol biodegradation. Process Biochem 39(8):983–988
Swetha VP, Phale PS (2005) Metabolism of carbaryl via 1, 2-dihydroxynaphthalene by soil isolates Pseudomonas sp. strains C4, C5, and C6. Appl Environ Microbiol 71(10):5951–5956
Hayatsu M, Tago K, Fukui M, Sekiya E (2005) Ecology of pesticide-degrading bacteria: degradation of organophosphorus and carbamate insecticides. ACS Publications, pp 82–91
Zhu S, Qiu J, Wang H, Wang X, Jin W, Zhang Y, Zhang C, Hu G, He J, Hong Q (2018) Cloning and expression of the carbaryl hydrolase gene mcbA and the identification of a key amino acid necessary for carbaryl hydrolysis. J Hazard Mater 344:1126–1135
Schirmer M, Ijaz UZ, D’Amore R, Hall N, Sloan WT, Quince C (2015) Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res 43(6):e37–e37
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genomics Proteomics Bioinform 13(5):278–289
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9(1):75
Cros M-J, De Monte A, Mariette J, Bardou P, Grenier-Boley B, Gautheret D, Touzet H, Gaspin C (2011) RNAspace.org: an integrated environment for the prediction, annotation, and analysis of ncRNA. RNA 17(11):1947–1956
Trivedi VD, Jangir PK, Sharma R, Phale PS (2016) Insights into functional and evolutionary analysis of carbaryl metabolic pathway from Pseudomonas sp. strain C5pp. Sci Rep 6:38430
Singh R, Trivedi VD, Phale PS (2013) Metabolic regulation and chromosomal localization of carbaryl degradation pathway in Pseudomonas sp. strains C4, C5 and C6. Arch Microbiol 195(8):521–535
Trivedi VD, Jangir PK, Sharma R (2016) Draft genome sequence of carbaryl-degrading soil isolate Pseudomonas sp. strain C5pp. Genome Announc 4(3):e00526–e00516
Fernández M, Niqui-Arroyo JL, Conde S, Ramos JL, Duque E (2012) Enhanced tolerance to naphthalene and enhanced rhizoremediation performance for Pseudomonas putida KT2440 via the NAH7 catabolic plasmid. Appl Environ Microbiol 78(15):5104–5110
Samanta SK, Singh OV, Jain RK (2002) Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol 20(6):243–248
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31670112, 31870092) and the National Key R&D Program of China (2017YFD0800702).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, S., Wang, H., Jiang, W. et al. Genome Analysis of Carbaryl-Degrading Strain Pseudomonas putida XWY-1. Curr Microbiol 76, 927–929 (2019). https://doi.org/10.1007/s00284-019-01637-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01637-4