Abstract
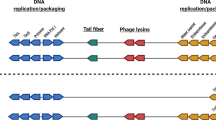
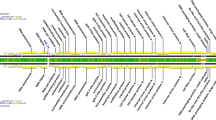
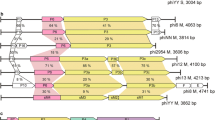
Bacteriophages remain an understudied component of bacterial communities. Therefore, our laboratory has initiated an effort to isolate large numbers of bacteriophages that infect Caulobacter crescentus to provide an estimate of the diversity of bacteriophages that infect this common environmental bacterium. The majority of the new isolates are phicbkviruses, a genus of giant viruses that appear to be Caulobacter specific. However, we have also isolated several Podoviruses with icosahedral heads and small tails. One of these Podoviruses, designated Lullwater, is similar to two previously isolated Caulobacter phages, Cd1 and Percy. All three have genomes that are approximately 45 kb and contain approximately 30 genes. The gene order is conserved among the three genomes with one of the genes coding for a DNA polymerase that has homology to the family of T7 DNA polymerases. Phylogenetic trees based on either the DNA polymerase or the RNA polymerase amino acid sequences suggests that the three phages represent a new branch of the T7virus tree. Based on these similarities, we concluded that Cd1, Lullwater, and Percy comprise a new group in the T7virus genus.




Similar content being viewed by others
References
Agabian-Keshishian N, Shapiro L (1970) Stalked bacteria: properties of deoxyribonucleic acid bacteriophage CbK. J Virol 5(6):798–800
Ahern SJ, Das M, Bhowmick TS, Young R, Gonzalez CF (2014) Characterization of novel virulent broad-host-range Phages of Xylella fastidiosa and Xanthomonas. J Bacteriol 196(2):459–471
Ash K, Drake KM, Gibbs WS, Ely B (2017) Genomic diversity of type B3 bacteriophages of Caulobacter crescentus. Curr Microbiol 74:779–786
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Darling ACE, Bau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
de Leeuw M, Baron M, Brenner A, Kushmaro A (2017) Genome analysis of a novel broad host range Proteobacteria Phage isolated from a bioreactor treating industrial wastewater. Genes (Basel) 8(1):40
Ely B, Croft RH (1982) Transposon mutagenesis of Caulobacter crescentus. J Bacteriol 149(2):620–625
Ely B, Johnson RC (1977) Generalized transduction in Caulobacter crescentus. Genetics 87:391–399
Ely B, Gibbs WS, Diez S, Ash K (2015) The Caulobacter crescentus transducing phage Cr30 is a unique member of the T4-like family of myophages. Curr Microbiol 70:854–858
Gill JJ, Berry JD, Russell WK, Lessor L, Escobar-Garcia DA, Hernandez D, Kane A, Keene J, Maddox M, Martin R, Mohan S, Thorn AM, Russell DH, Young R (2012) The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13:542
Hamdi S, Rousseau GM, Labrie SJ, Kourda RS, Tremblay DM, Moineau S, Slama KB (2016) Characterization of five Podoviridae Phages infecting Citrobacter freundii. Front Microbiol 7:1023. https://doi.org/10.3389/fmicb.2016.01023
Johnson RC, Ely B (1977) Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25–32
Johnson RC, Wood NB, Ely B (1977) Isolation and characterization of bacteriophages for Caulobacter crescentus. J Gen Virol 37:323–335
Lavigne R, Burkal’tseva MV, Robben J, Sykilinda NN, Kurochkina LP, Grymonprez B, Jonckx B, Krylov VN, Mesyanzhinov VV, Volckaert G (2003) The genome of bacteriophage φKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 312(1):49–59
Lerma R, Tidwell TJ, Cahill JL, Rasche ES, Kuty Everett GF (2015) Complete genome sequence of Caulobacter crescentus Podophage Percy. Genome Announc 3(6):e01373-15
Papadopoulos JS1, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23(9):1073–1079
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945
Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ (2004) Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J Mol Biol 335(5):1151–1171
West D, Lagenaur C, Agabian N (1976) Isolation and characterization of Caulobacter crescentus bacteriophage phiCd1. J Virol 17(2):568–575
Acknowledgements
The authors would like to thank Kiesha Wilson for her advice and assistance. This work was funded in part by NIH grant R25GM076277.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors state that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nguyen, D., Ely, B. A Genome Comparison of T7-like Podoviruses That Infect Caulobacter crescentus. Curr Microbiol 75, 760–765 (2018). https://doi.org/10.1007/s00284-018-1445-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1445-9