Abstract
This review aimed to investigate the role of Helicobacter pylori flagella on the pathogenicity of this bacterium in humans. Helicobacter pylori is a flagellated pathogen that colonizes the human gastroduodenal mucosa and produces inflammation, and is responsible for gastrointestinal disease. Its pathogenesis is attributed to colonization and virulence factors. The primary function of H. pylori flagella is to provide motility. We believe that H. pylori flagella play an important role in the colonization of the gastrointestinal mucosa. Therefore, we reviewed previous studies on flagellar morphology and motility in order to explore the relationship between H. pylori flagella and pathogenicity. Further investigation is required to confirm the association between flagella and pathogenicity in H. pylori.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
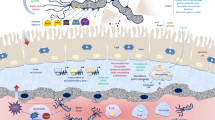
Helicobacter pylori is a flagellated microaerophilic gram-negative bacillus that is known to colonize the gastrointestinal mucosa of almost half the global human population with varying prevalence rates across different geographical regions [13]. Helicobacter pylori is perhaps the most infectious of all known bacteria. Although some believe that H. pylori is a type of “commensal bacterium” [52], it cannot be classified as normal flora because all patients with gastroduodenal H. pylori colonization show histological gastroenteritis [39], which can develop into a number of gastric diseases such as chronic gastritis, duodenitis, peptic ulcers (gastric and duodenal), mucosa-associated lymphoid tissue (MALT), atrophic gastritis, and gastric adenocarcinoma. This bacterium exhibits allelic diversity and genetic variability. Thus, infection might appear as a high rate of mixed infections, indicating that one person might be infected with multiple strains of H. pylori. The mixed infection rate is high in epidemic areas with a high incidence [35]. The pathogenesis of H. pylori infection is partly dependent on colonization and virulence factors [39], and flagella play an important role in the colonization of the gastrointestinal mucosa [24]. The difference (heterogeneity) in the motilities of colonizing strains was first reported by [17], but failed to attract much attention. The role of heterogeneity in the motility of H. pylori has not been sufficiently explored.
Morphology and Structure of H. pylori Flagella
The bacterial flagellum is a complex motility organ composed of multiple types of protein subunits [46]. Each flagellum consists of three components [44, 53]: the basal body, hook, and filament. Electron microscopic observation of the H. pylori flagellum reveals the presence of a sheath and a terminal bulb [68]. The function of the membrane-like flagellar sheath of H. pylori is hitherto unknown, and little is known of its composition [47]. Helicobacter pylori has 4–8 unipolar flagella [42]; however, it remains controversial whether the flagella are uni- or bipolar [25]. Table 1 presents the structural composition and functions of H. pylori flagellar structures.
Functions and Pathogenicity of H. pylori Flagella
Helicobacter pylori flagella produce different types of motility [24], including “swimming motility,” “spreading motility,” and “swarming motility,” which are defined as movement in liquid media, movement in soft agar (0.3% agar concentration), and movement on the surface of semi-solid or solid media, respectively. Helicobacter pylori flagella may influence their colonization in bacteria, inflammation, and immune evasion.
Colonization and Colonization sites
Colonization
The viability of H. pylori on the surface of the gastrointestinal mucosa depends on its colonization factors such as urease, motility, chemotaxis, outer membrane proteins, and the special helix morphology of the bacterium [15, 44, 53, 78, 85]. The optimal pH of H. pylori is neutral [28, 76], while the optimal pH of H. pylori in liquid media is 8.5 (slightly alkaline) [4]. Thus, the acidic environment of the stomach is not suitable for its growth. It is generally believed that urease plays a protective role in H. pylori [74], facilitating colonization of the mucosa of the stomach by hydrolyzing urea into NH3 and CO2 [69]. The NH3 produced by the action of urease neutralizes stomach acid and increases the pH of the surrounding cells [15]. In addition, urease participates in the inflammatory reaction and facilitates adhesion by interacting with the CD74 receptor on gastric epithelial cells [48, 50] isolated urease-negative H. pylori mutants from patients with peptic ulcers, and found that this strain successfully colonized the stomach of Mongolian gerbils and caused ulcers; therefore, the role of urease as a colonization factor is uncertain. The importance of motility as a colonization factor in H. pylori was first demonstrated in the study by [17], who demonstrated that germ-free piglets exhibited a higher infection rate when infected with motile H. pylori than when infected with the non-motile strain; moreover, the motile strain also colonized the stomachs for a longer duration in the stomach of germ-free piglets. Similarly, many animal studies using motility-deficient mutants, including the H. pylori motB mutant [62], fliD mutant [37], putA mutant [56], and a chemotaxis mutant strain [49, 56], have shown similar findings. The spreading motility of these mutant strains is weak and their colonization in the stomach of the animals was reduced. The most convincing evidence for the role of motility in H. pylori colonization came from the study by [5, 60] found that changes in flagellin glycosylation affected strain motility. When the glycosylation level of the FlaA protein was increased, the strain motility and colonization load both increased. These studies have convincingly indicated that flagellar motility is an important factor influencing colonization. The colonization of H. pylori is expressed as colonization density or load. The number of colonies per gram of gastric mucosa (CFU/g) [6, 49] is determined by quantitative culture. Helicobacter pylori DNA can be quantitatively measured by polymerase chain reaction (PCR) [20].
Motility is measured by three different methods. First, swimming motility can be directly determined by the average swimming velocity of bacteria in the gastric mucosal layer using phase-contrast microscopy [8]. Second, spreading motility can be determined by assessing the growth ring diameter in semi-solid agar using puncture inoculation [49, 60, 62]. Third, swarming motility can be determined by examining the growth ring diameter on the surface of semi-solid medium [51, 56] using quantitative inoculation. These studies showed that the H. pylori colonization load in gastric mucosa was positively correlated with motility in animal infection models.
Colonization Site
Colonization by H. pylori is not evenly distributed. Colonization in the stomach is usually observed at the gastric antrum [51]. However, H. pylori colonies can also be found in other sites. For example, H. pylori has been known to colonize the duodenum and is recognized as the primary cause of idiopathic duodenal ulcers [65, 66]. Although the incidence is low (6.9%), it can be assumed that the strains colonizing different gastrointestinal sites might have different origins, because H. pylori infection in the stomach has been demonstrated to be heterogeneous [35]. Helicobacter pylori may also colonize the colon [57, 58, 81], but its origin remains unclear.
Immune Inflammation and Evasion
The motility and colonization load of H. pylori are positively correlated with neutrophilin filtration [1, 34, 49]. Colonization is the basis of the inflammatory reaction induced by H. pylori, and motility is a critical colonization determinant that affects the infection outcome. Furthermore, the flagellum also influences inflammation and immune evasion.
Immune Inflammation
The main structural proteins of the H. pylori flagellum include HpaA, FlaA, FlaB, FliD, and FlgK. Of these, HpaA, FlaA, and FlaB have been found to be expressed in H. pylori strains isolated from biopsy specimens of patients with stomach disease. These flagellins are the primary targets of the humoral immunity after infection, and induce antibody responses [79]. In contrast, an important study showed that FlaA was antigenic but not immunogenic [72]. The role of H. pylori flagellins in immune inflammation is yet unknown.
Immune Evasion
Helicobacter pylori infections usually occur during childhood and last for a lifetime if left untreated with antibiotics [45]. The human immune system cannot eliminate these bacteria primarily because of the bacterial ability of immune evasion. Although H. pylori flagellin can induce anti-flagellin antibodies in infected patients, it is not recognized by toll-like receptor 5 (TLR5), a member of the toll-like receptor family, which is activated by most bacterial flagellins [1, 54]. One possible reason may be that the flagellins, especially FlaA, are not exposed, and thus cannot be detected in the infected gastric epithelial cells [22]. Other bacterial flagella induce interleukin 8 (IL-8) secretion, leading to an inflammatory reaction. However, H. pylori flagellin does not typically induce IL-8 secretion in gastric epithelial cells. Although highly motile strains of H. pylori have been shown to elicit a higher level of IL-8 production [42], the flagellar sheath HpaA probably shields the flagellin from recognition by TLR5 [10].
Relationship Between H. pylori Flagellar Structure, Motility, Chemotaxis, and Colonization
In the flagellar structure, the C ring complex is composed of FliM, FliN, FliY, and FliG. Typically, fliM, fliY, and fliG mutant H. pylori strains cannot produce flagella. Although the fliN mutant strain can produce flagella, they are “paralyzed” and unable to move [46], resulting in a non-motile bacterium. In the flagellar structure, the “motor” is important for bacterial motility. motB-deficient H. pylori have been reported to exhibit normal flagellar structure but no motility, and the colonization load of this strain is significantly lower than controls containing motB in infected mice [62]. fliF, fliS, flhB, fliQ, fliG, or fliI mutant strains did not produce any flagella and were non-motile, while the flhA mutant strain produced short flagella [3]. FlgE is the main protein of the flagellar hook, and strains lacking the flgE gene expectedly showed no motility [61]. FlaA and FlaB are the components of the flagellar filament and are important for motility. Strains lacking the flaA and flaB genes exhibit reduced irregular flagella and lower motility. The flaA and flaB double-mutant strain is completely non-motile. The flaA and flaB mutant strains have reduced colonizing ability [18, 32], and cannot colonize even with a longer period of incubation in animal models [18]. Mutation of the flagellar filament fliD gene results in non-motile bacteria with short flagella, and this strain is unable to colonize the gastric mucosa of mice [37]. The FaaA protein is required for flagellar and proper flagellar localization as well as for optimal flagellar function. This protein is exported to the outer membrane and subsequently becomes a component of the flagellar sheath. Helicobacter pylori mutant strains deficient in faaA exhibited decreased motility and less efficient colonization of the stomach in mice compared to the wild-type H. pylori strain at the early stages of infection [10, 68].
Flagellar hook substructure reaches its optimal length sensed by the ‘checkpoint control’ protein FliK, export of the anti-sigma factor FlgM is triggered releasing σ28 from a σ28-FlgM complex which in turn allows the subsequent expression of σ28 dependent genes. In fliK, mutants hook to filament transition do not occur and long hooks of unregulated length termed polyhooks are formed [55]. It is demonstrated that FliK is necessary for upregulation of cagA. Expression and flagellar regulatory system of H. pylori is directly required for upregulation of the major virulence gene cagA in gastric cell associated H. pylori.
The direction of flagellar rotation is cooperatively controlled by the chemotaxis-signaling protein CheY and the flagellar rotor protein FliN [44]. Strains with mutant chemotaxis genes cheW, cheV [67], cheY, cheA [8], and tlpB [49] have less motility and reduced colonization load [49, 80]. In contrast, a study by Williams et al. demonstrated that chemotaxis gene mutants (ΔcheY, ΔcheW) displayed an adequate colonization load but a reduced inflammatory response [83]. Reference [30] investigated a new protein, ChePep, located in the flagellar pole, which regulates flagellar rotation and controls H. pylori chemotaxis. Strains with the ChePep gene mutation exhibited reduced flagellar motility.
The only known phosphatase in H. pylori is CheZ, called CheZHP in this system. It is reported that CheZHP localization depends on the ChePep chemotaxis protein [29] and conversely ChePep localization depends on CheZHP, which raises the intriguing possibility that some phosphatases, including CheZHP and ChePep, exist in a complex that is distinct from the core chemotaxis signaling and flagellar complexes [43].
Reference [70] demonstrated that the colonization of a TlpD-controlled chemotaxis gene mutant strain in the gastric antrum was significantly reduced. Similarly, it was found that the colonization of strains with chemotaxis gene mutants (ΔcheY) and especially the motility gene mutants (ΔmotB) was reduced [1]. Future studies should clarify the relationship between chemotaxis and colonization.
All members of the Epsilonproteobacteria have their flagella located at either one or both cellular poles [2, 27, 40, 64, 73, 75, 84]. Campylobacter jejuni and Helicobacter pylori are the most studied epsilonproteobacteria because they are important human pathogens. In addition to their unique structural features revealed by cryoelectron tomography [11], Campylobacter and Helicobacter flagella exhibit unique aspects in the regulation of the expression of their flagellar genes and in the assembly of their flagellar structure [23, 44]. Regulation of flagellar gene expression in Campylobacter and Helicobacter is also unique, involving a two-component system (FlgRS), the FlhF GTPase, and the transcription factors σ54 and σ28 [7, 33, 63].
Discussion
Although many components of the H. pylori lagella have been characterized and data regarding flagellar function and regulation are rapidly increasing, certain aspects of the H. pylori system, in particular those that differ from the well-studied model systems, are still poorly understood and require further investigation. These regulatory mechanisms appear to act at the bottom of the putative transcriptional hierarchy that governs flagellar biosynthesis in H. pylori. In contrast, the mechanisms at the top of the hierarchy that actually trigger the initiation of flagellar gene transcription are completely unknown.
Previous studies have explored the spreading and swarming motilities; however, whether these two types of motility are equivalent remains unclear.
Adhesion is an important factor that mediates the pathogenic role of bacterial flagellum [19, 26]. However, unlike other bacteria, H. pylori adhesion on gastric epithelial cells is not dependent on flagellin [12] and is not influenced by reduction in bacterial flagella. Moreover, it is related to mutations of flagellar genes. For example, the adhesion ability of a flaA::cat/flab::km mutant strain without flagella is adequate while that of the flbA mutant strain is significantly reduced.
Exploring the relationship between H. pylori flagellar motility and gastrointestinal mucosa colonization can facilitate the understanding of H. pylori pathogenesis, especially the heterogeneity of motility in mixed infections, and needs to be further investigated.
References
Aihara E, Closson C, Matthis AL, Schumacher MA, Engevik AC, Zavros Y, Ottemann KM, Montrose MH (2014) Motility and chemotaxis mediate the preferential colonization of gastric injury sites by Helicobacter pylori. PLoS Pathog 10(7):e1004275
Alain K, Querellou J, Lesongeur F, Pignet P, Crassous P, Raguénès G, Cueff V, Cambon-Bonavita MA (2002) Caminibacter hydrogeniphilus gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing bacterium isolated from an East Pacific Rise hydrothermal vent. Int J Syst Evol Microbiol 52:1317–1323
Allan E, Dorrell N, Foynes S, Anyim M, Wren BW (2000) Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin A biosynthesis. J Bacteriol 182(18):5274–5277
Andersen AP, Elliott DA, Lawson M, Barland P, Hatcher VB, Puszkin EG (1997) Growth and morphological transformations of Helicobacter pylori in broth media. J Clin Microbiol 35(11):2918–2922
Asakura H, Churin Y, Bauer B, Boettcher JP, Bartfeld S, Hashii N, Kawasaki N, Mollenkopf HJ, Jungblut PR, Brinkmann V, Meyer TF (2010) Helicobacter pylori HP0518 affects flagellin glycosylation to alter bacterial motility. Mol Microbiol 78(5):1130–1144
Atherton JC, Tham KT, Peek RM Jr, Cover TL, Blaser MJ (1996) Density of Helicobacter pylori infection in vivo as assessed by quantitative culture and histology. J Infect Dis 174(3):552–556
Balaban M, Hendrixson DR (2011) Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog 7:e1002420
Bansil R, Celli JP, Hardcastle JM, Turner BS (2013) The influence of mucus microstructure and rheology in Helicobacter pylori infection. Front Immunol 4:310
Basso D, Plebani M, Kusters JG (2010) Pathogenesis of Helicobacter pylori infection. Helicobacter 15(Suppl 1):14–20
Carlsohn E, Nystrom J, Bolin I, Nilsson CL, Svennerholm AM (2006) HpaA is essential for Helicobacter pylori colonization in mice. Infect Immun 74(2):920–926
Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Müller A, Dobro MJ, Jensen GJ (2011) Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981
Clyne M, Ocroinin T, Suerbaum S, Josenhans C, Drumm B (2000) Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect Immun 68(7):4335–4339
Cover TL, Blaser MJ (2009) Helicobacter pylori in health and disease. Gastroenterology 136(6):1863–1873
Douillard FP, Ryan KA, Hinds J, O’Toole PW (2009) Effect of FliK mutation on the transcriptional activity of the σ54 sigma factor RpoN in Helicobacter pylori. Microbiology 155(Pt 6):1901–1911
Dunne C, Dolan B, Clyne M (2014) Factors that mediate colonization of the human stomach by Helicobacter pylori. World J Gastroenterol 20(19):5610–5624
Dyer CM, Vartanian AS, Zhou H, Dahlquist FW (2009) A molecular mechanism of bacterial flagellar motor switching. J Mol Biol 388(1):71–84
Eaton KA, Morgan DR, Krakowka S (1992) Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol 37(2):123–127
Eaton KA, Suerbaum S, Josenhans C, Krakowka S (1996) Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun 64(7):2445–2448
Friedlander RS, Vlamakis H, Kim P, Khan M, Kolter R, Aizenberg J (2013) Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc Natl Acad Sci USA 110(14):5624–5629
Furuta T, Kaneko E, Suzuki M, Arai H, Futami H (1996) Quantitative study of Helicobacter pylori in gastric mucus by competitive PCR using synthetic DNA fragments. J Clin Microbiol 34(10):2421–2425
Geis G, Suerbaum S, Forsthoff B, Leying H, Opferkuch W (1993) Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J Med Microbiol 38(5):371–377
Gewirtz AT, Yu Y, Krishna US, Israel DA, Lyons SL, Peek RM Jr (2004) Helicobacter pylori flagellin evades toll-like receptor 5-mediated innate immunity. J Infect Dis 189(10):1914–1920
Gilbreath JJ, Cody WL, Merrell DS, Hendrixson DR (2011) Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol Mol Biol Rev 75:84–132
Gu HY (2008) The study on the mechanisms of Helicobacter pylori motility in gastric mucosal colonization. Chin J Lab Med 31:733–736 (Chinese)
Gu HY (2013) Bacterial flagella staining technology and its application. Chin J Microbiol Immunol 33:462–464 (Chinese)
Haiko J, Westerlund-Wikstrom B (2013) The role of the bacterial flagellum in adhesion and virulence. Biology 2(4):1242–1267 (Basel)
Han C, Kotsyurbenko O, Chertkov O, Held B, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Cheng JF, Tapia R, Goodwin LA, Pitluck S, Liolios K, Pagani I, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Brambilla EM, Rohde M, Spring S, Sikorski J, Goker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Detter JC (2012) Complete genome sequence of the sulfur compounds oxidizing chemolithoautotroph Sulfuricurvum kujiense type strain (YK-1(T)). Stand Genom Sci 6:94–103
Ho B, Vijayakumari S (1993) A simple and efficient continuous culture system for Helicobacter pylori. Microbios 76(306):59–66
Howitt, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva (2001) ChePep controls Helicobacter pylori Infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2:e00098-11
Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR (2011) ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. MBio 2(4):e00098-11
Ibuki T, Uchida Y, Hironaka Y, Namba K, Imada K, Minamino T (2013) Interaction between FliJ and FlhA, components of the bacterial flagellar type III export apparatus. J Bacteriol 195(3):466–473
Josenhans C, Labigne A, Suerbaum S (1995) Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol 177(11):3010–3020
Joslin SN, Hendrixson DR (2009) Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol 191:2656–2667
Kao CY, Sheu BS, Sheu SM, Yang HB, Chang WL, Cheng HC, Wu JJ (2012) Higher motility enhances bacterial density and inflammatory response in dyspeptic patients infected with Helicobacter pylori. Helicobacter 17(6):411–416
Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, Falush D, Suerbaum S (2011) Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA 108(12):5033–5038. doi:10.1073/pnas.1018444108
Khalifeh Gholi M, Kalali B, Formichella L, Gottner G, Shamsipour F, Zarnani AH, Hosseini M, Busch DH, Shirazi MH, Gerhard M (2013) Helicobacter pylori FliD protein is a highly sensitive and specific marker for serologic diagnosis of H. pylori infection. Int J Med Microbiol 303(8):618–623
Kim JS, Chang JH, Chung SI, Yum JS (1999) Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol 181(22):6969–6976
Kinoshita M, Hara N, Imada K, Namba K, Minamino T (2013) Interactions of bacterial flagellar chaperone-substrate complexes with FlhA contribute to co-ordinating assembly of the flagellar filament. Mol Microbiol 90(6):1249–1261
Kusters JG, van Vliet AH, Kuipers EJ (2006) Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19(3):449–490
Labrenz M, Grote J, Mammitzsch K, Boschker HT, Laue M, Jost G, Glaubitz S, Jürgens K (2013) Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic Baltic Sea redoxcline, and an emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol 63:4141–4148
Lam WW, Woo EJ, Kotaka M, Tam WK, Leung YC, Ling TK, Au SW (2010) Molecular interaction of flagellar export chaperone FliS and cochaperone HP1076 in Helicobacter pylori. FASEB J 24(10):4020–4032
Lawson AJ (2011) Helicobacter. In: James V, Karen C, Guido F (eds) Manual of clinical microbiology, 10th edn. ASM, Washington, pp 900–915
Lertsethtakarn P, Howitt MR, Castellon J, Amieva MR, Ottemann KM (2015) Helicobacter pylori CheZ(HP) and ChePep form a novel chemotaxis-regulatory complex distinct from the corechemotaxis signaling proteins and the flagellar motor. Mol Microbiol 97(6):1063–1078
Lertsethtakarn P, Ottemann KM, Hendrixson DR (2011) Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410
Lina TT, Alzahrani S, Gonzalez J, Pinchuk IV, Beswick EJ, Reyes VE (2014) Immune evasion strategies used by Helicobacter pylori. World J Gastroenterol 20(36):12753–12766
Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM (2009) Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol 191(23):7147–7156
Luke CJ, Penn CW (1995) Identification of a 29 kDa flagellar sheath protein in Helicobacter pylori using a murine monoclonal antibody. Microbiology 141(Pt 3):597–604
Mascellino MT, Margani M, Oliva A (2009) Helicobacter pylori: determinant and markers of virulence. Dis Markers 27(3):137–156
McGee DJ, Langford ML, Watson EL, Carter JE, Chen YT, Ottemann KM (2005) Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun 73(3):1820–1827
Mine T, Muraoka H, Saika T, Kobayashi I (2005) Characteristics of a clinical isolate of urease-negative Helicobacter pylori and its ability to induce gastric ulcers in Mongolian gerbils. Helicobacter 10(2):125–131
Mishra J, Panigrahi S (2011) A study of changes in stomach wall at sites other than the ulcer in chronic duodenal ulcer patients. Indian J Surg 73(4):262–263
Mishra S (2013) Is Helicobacter pylori good or bad? Eur J Clin Microbiol Infect Dis 32(3):301–304
Mobley HT, Mendz GL, Hazell SL (2001) Motility, chemotaxis, and flagella. Helicobacter pylori: physiology and genetics. ASM, Washington
Mori J, Vranac T, Smrekar B, Cernilec M, Serbec VC, Horvat S, Ihan A, Bencina M, Jerala R (2012) Chimeric flagellin as the self-adjuvanting antigen for the activation of immune response against Helicobacter pylori. Vaccine 30(40):5856–5863
Muramoto K, Makishima S, Aizawa SI, Macnab RM (1998) Effect of cellular level of FliK on flagellar hook and filament assembly in Salmonella typhimurium. J Mol Biol 277:871–882
Nakajima K, Inatsu S, Mizote T, Nagata Y, Aoyama K, Fukuda Y, Nagata K (2008) Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res 29(1):9–18
Oliveira AG, das Sanna MGP, Rocha GA, Rocha AM, Santos A, Dani R, Marinho FP, Moreira LS, de Ferrari MLA, Moura SB, Castro LP, Queiroz DM (2004) Helicobacter species in the intestinal mucosa of patients with ulcerative colitis. J Clin Microbiol 42(1):384–386
Oliveira AG, Rocha GA, Rocha AM, Sanna M, Moura SB, Dani R, Marinho FP, Moreira LS, Ferrari Mde L, Castro LP, Queiroz DM (2006) Isolation of Helicobacter pylori from the intestinal mucosa of patients with Crohn’s disease. Helicobacter 11(1):2–9
O’Neill J, Xie M, Hijnen M, Roujeinikova A (2011) Role of the MotB linker in the assembly and activation of the bacterial flagellar motor. Acta Crystallogr D Biol Crystallogr 67(Pt 12):1009–1016
Osaki T, Hanawa T, Manzoku T, Fukuda M, Kawakami H, Suzuki H, Yamaguchi H, Yan X, Taguchi H, Kurata S, Kamiya S (2006) Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J Med Microbiol 55(Pt 11):1477–1485
O’Toole PW, Kostrzynska M, Trust TJ (1994) Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol Microbiol 14(4):691–703
Ottemann KM, Lowenthal AC (2002) Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun 70(4):1984–1990
Pereira L, Hoover TR (2005) Stable accumulation of sigma54 in Helicobacter pylori requires the novel protein HP0958. J Bacteriol 187:4463–4469
Pérez-Rodríguez I, Ricci J, Voordeckers JW, Starovoytov V, Vetriani C (2010) Nautilia nitratireducens sp. nov., a thermophilic, anaerobic, chemosynthetic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol 60:1182–1186
Pietroiusti A, Forlini A, Magrini A, Galante A, Bergamaschi A (2008) Isolated Helicobacter pylori duodenal colonization and idiopathic duodenal ulcers. Am J Gastroenterol 103(1):55–61
Pietroiusti A, Luzzi I, Gomez MJ, Magrini A, Bergamaschi A, Forlini A, Galante A (2005) Helicobacter pylori duodenal colonization is a strong risk factor for the development of duodenal ulcer. Aliment Pharmacol Ther 21(7):909–915
Pittman MS, Goodwin M, Kelly DJ (2001) Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147(Pt 9):2493–2504
Radin JN, Gaddy JA, Gonzalez-Rivera C, Loh JT, Algood HM, Cover TL (2013) Flagellar localization of a Helicobacter pylori autotransporter protein. MBio 4(2):e00613-12
Roesler BM, Rabelo-Goncalves EM, Zeitune JM (2014) Virulence Factors of Helicobacter pylori: a review. Clin Med Insights Gastroenterol 7:9–17
Rolig AS, Shanks J, Carter JE, Ottemann KM (2012) Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect Immun 80(10):3713–3720
Ryan KA, Karim N, Worku M, Penn CW, O’Toole PW (2005) Helicobacter pylori flagellar hook-filament transition is controlled by a FliK functional homolog encoded by the gene HP0906. J Bacteriol 187(16):5742–5750
Sanders CJ, Yu Y, Moore DA 3rd, Williams IR, Gewirtz AT (2006) Humoral immune response to flagellin requires T cells and activation of innate immunity. J Immunol 177(5):2810–2818
Schuster SC, Baeuerlein E (1992) Location of the basal disk and a ringlike cytoplasmic structure, two additional structures of the flagellar apparatus of Wolinella succinogenes. J Bacteriol 174:263–268
Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G (1998) The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114(1):58–70
Sikorski J, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, Saunders E, Bruce D, Goodwin L, Pitluck S, Ovchinnikova G, Pati A, Ivanova N, Mavromatis K, Chen A, Palaniappan K, Chain P, Land M, Hauser L, Chang YJ, Jeffries CD, Brettin T, Detter JC, Han C, Rohde M, Lang E, Spring S, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP (2010) Complete genome sequence of Sulfurospirillum deleyianum type strain (5175). Stand Genom Sci 2:149–157
Slonczewski JL, McGee DJ, Phillips J, Kirkpatrick C, Mobley HL (2000) pH-dependent protein profiles of Helicobacter pylori analyzed by two-dimensional gels. Helicobacter 5(4):240–247
Smith TG, Pereira L, Hoover TR (2009) Helicobacter pylori FlhB processing-deficient variants affect flagellar assembly but not flagellar gene expression. Microbiology 155(Pt 4):1170–1180
Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR (2012) Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog 8(3):e1002603
Tang RX, Luo DJ, Sun AH, Yan J (2008) Diversity of Helicobacter pylori isolates in expression of antigens and induction of antibodies. World J Gastroenterol 14(30):4816–4822
Terry K, Williams SM, Connolly L, Ottemann KM (2005) Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73(2):803–811
Thomson JM, Hansen R, Berry SH, Hope ME, Murray GI, Mukhopadhya I, McLean MH, Shen Z, Fox JG, El-Omar E, Hold GL (2011) Enterohepatic helicobacter in ulcerative colitis: potential pathogenic entities? PLoS ONE 6(2):e17184. doi:10.1371/journal.pone.0017184
Tsang J, Smith TG, Pereira LE, Hoover TR (2013) Insertion mutations in Helicobacter pylori flhA reveal strain differences in RpoN-dependent gene expression. Microbiology 159(Pt 1):58–67
Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM (2007) Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun 75(8):3747–3757
Wirsen CO, Sievert SM, Cavanaugh CM, Molyneaux SJ, Ahmad A, Taylor LT, DeLong EF, Taylor CD (2002) Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl Environ Microbiol 68:316–325
Zhang J, Qian J, Zhang X, Zou Q (2014) Outer membrane inflammatory protein A, a new virulence factor involved in the pathogenesis of Helicobacter pylori. Mol Biol Rep 41(12):7807–7814
Acknowledgements
This study was funded by the Zhejiang Provincial Natural Science Foundation (LZ14H200001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no any conflict of interests.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gu, H. Role of Flagella in the Pathogenesis of Helicobacter pylori . Curr Microbiol 74, 863–869 (2017). https://doi.org/10.1007/s00284-017-1256-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1256-4