Abstract
Streptococcus dysgalactiae is a pyogenic species pathogenic both for humans and animals. Until recently, it has been considered an exclusive animal pathogen causing infections in wild as well as domestic animals. Currently, human infections are being reported with increasing frequency, and their clinical picture is often similar to the ones caused by Streptococcus pyogenes. Due to the fact that S. dysgalactiae is a heterogeneous species, it was divided into two subspecies: S. dysgalactiae subsp. equisimilis (SDSE) and S. dysgalactiae subsp. dysgalactiae (SDSD). The first differentiation criterion, described in 1996, was based on strain isolation source. Currently applied criteria, published in 1998, are based on hemolysis type and Lancefield group classification. In this study, we compared subspecies identification results for 36 strains isolated from clinical cases both in humans and animals. Species differentiation was based on two previously described criteria as well as MALDI-TOF and genetic analyses: RISA and 16S rRNA genes sequencing. Antimicrobial susceptibility profiles were also determined according to CLSI guidelines. The results presented in our study suggest that the subspecies differentiation criteria previously described in the above two literature positions seem to be inaccurate in analyzed group of strains, the hemolysis type on blood agar, and Lancefield classification should not be here longer considered as criteria in subspecies identification. The antimicrobial susceptibility tests indicate emerging of multiresistant human SDSE strains resistant also to vancomycin, linezolid and tigecycline, which might pose a substantial problem in treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptococcus dysgalactiae is a pyogenic species pathogenic both for humans and animals. Until recently, it has been considered an exclusive animal pathogen causing infections in wild animals, livestock, and pets. Currently, human infections are being reported with increasing frequency. Their clinical picture is often similar to the ones caused by group A streptococcus (Streptococcus pyogenes) [3].
S. dysgalactiae proved to be a heterogeneous species, what encouraged researchers to divide it into two subspecies: S. dysgalactiae subsp. equisimilis (SDSE) and S. dysgalactiae subsp. dysgalactiae (SDSD). The first differentiation criterion was proposed in 1996 by Vandamme et al. [22], second criteria—only 2 years later—in 1998 by Vieira et al. [23] which are being applied up to the present. Over the years, controversies arose over this classification, showing that the streptococci taxonomy has not been completed yet and generally used S. dysgalactiae subspecies division causes more questions than solutions. In some research reports, subspecies names are used instead of species (i.e., S. equisimilis), which causes unnecessary confusion in diagnostics [4, 19].
The presented study provides a new view on S. dysgalactiae identification, subspecies differentiation criteria and similarities between strains isolated from human and animal clinical materials.
Materials and Methods
Bacterial Strains
30 human S. dysgalactiae isolates from clinical cases (mainly wounds, abscesses, bedsores, noses, ears, throats) were obtained from Synevo Medical Laboratory in Łódź, Poland.
6 animal S. dysgalactiae isolates from clinical cases in pets (dogs—from wounds, abscesses, birth canal) were obtained from VETCOMPLEX Veterinary Diagnostic Centre in Łódź, Poland.
Phenotypic Bacterial Identification According to Vieira et al. [23]
Hemolysis type was evaluated as well as Lancefield group. The type of hemolysis was observed after a 24-h incubation at 37 °C (aerobic atmosphere) on agar plates containing 5 % defibrinated sheep blood. The type of surface C-polysaccharide, classifying analyzed strains to serological groups according to Lancefield, was evaluated using latex agglutination tests with Prolex Streptococcal Grouping Latex Kit.
Bacterial Identification Based on Cell Proteins Structure and DNA Sequences
MALDI-TOF technique (matrix-assisted laser desorption ionization-time of flight) [5] which compares cell proteins specters with database (bioMérieux VITEK® MS) was employed to identify the analyzed clinical strains.
All bacterial strains were also identified by means of RISA (16S–23S rDNA intergenic spacer region) method. PCR reactions were conducted using S. dysgalactiae species-specific primers (Fwd 5′-TGGAACACGTTAGGGTCG-3′, Rev 5′-CTTAACTAGAAAAACTCTTGATTATTC-3′) [9] and SDSE subspecies-specific primers (Fwd 5′-GGTACTGTTGAGGGGACGAA-3′, Rev 5′-CGATTGAGGAGTCACGTTCA-3′) [16].
Genetic identification of the selected strains was also confirmed by means of amplification of 16S rRNA coding genes from the selected strains, using primers described in the paper (8F 5′-GAGAGTTTGATCCTGGCTCAG-3′, 1942R 5′-TACGGCTACCTTGTTACGACT-3′) [21]. Afterward, amplified DNA fragments were sequenced and compared with Genbank database. The obtained sequences were published in Genbank.
Antimicrobial Susceptibility
The antibiotic susceptibility tests of the analyzed strains were performed by means of disk diffusion method according to Clinical & Laboratory Standards Institute (CLSI) [6] and Polish National Reference Centre for Antibiotics (KORLD) [26] guidelines. Susceptibility to 10 antibiotics, representative of the main antibiotic classes and recommended for streptococcal infections treatment, were determined: penicillin (10 U), erythromycin (15 µg), clindamycin (2 µg), ofloxacin (5 µg), levofloxacin (5 µg), vancomycin (30 µg), tetracycline (30 µg), tigecycline (15 µg), linezolid (30 µg), and cefotaxime (30 µg). The results were interpreted according to CLSI recommendations [6].
Results
Our findings refer to the strains collected over the period of 2014 in the area of Łódź. They were more frequently isolated from purulent infections in humans than in animals.
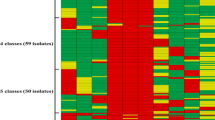
The analyzed S. dysgalactiae strains were assigned to subspecies according to the criteria proposed by Vandamme et al., Vieira et al., as well as to the results of MALDI-TOF method and genetic studies (Table 1). The classification by Vandamme et al. was based on strain isolation sources. 30 human isolates were classified as SDSE, whereas 6 animal isolates were classified as SDSD subspecies.
Subspecies identification according to Vieira et al. was based on phenotypic characteristics: hemolysis type and Lancefield group. Among 30 human S. dysgalactiae isolates, nine (30 %) demonstrated α-type hemolysis, whereas 21 (70 %) demonstrated β-type hemolysis on blood agar. Among animal isolates, one (17 %) showed α-type hemolysis, whereas other five (83 %) showed β-type hemolysis on blood agar. Both human and animal isolates were classified to Lancefield groups C or G. 16 (53 %) human isolates belonged to group C, 14 (47 %) to group G. Among animal isolates, 1 (17 %) was classified to group C, whereas other 5 (83 %) to Lancefield group G. According to subspecies division criteria proposed by Vieira et al., among human isolates, 21 (70 %) might be classified to SDSE, 5 (17 %) to SDSD, but 4 isolates (13 %) did not match the criteria for neither of the subspecies. 5 (83 %) animal isolates might be classified to SDSE, whereas 1 (17 %) did not match the criteria for neither of the subspecies according to Vieira et al.
All analyzed strains were unequivocally identified as SDSE by means of MALDI-TOF technique and RISA genetic method. Additionally, sequencing of the genes coding for 16S rRNA, isolated from seven strains exhibiting various phenotypic characteristics (Table 1, bolded strains) confirmed their phylogenetic affiliation to SDSE subspecies. All obtained sequences showed over 99 % similarity to SDSE sequences previously deposited in Genbank. Sequences of genes coding for 16S rRNA obtained from six human isolates were also deposited in Genbank under accession numbers KR092084, KR559932, KR559933, KR559934, KR559935, and KR559936, whereas one from animal strain isolated from a dog was deposited in KT232322.
Antimicrobial Susceptibility
Antibiotic susceptibility profiles of human and animal isolates were roughly similar. The analyzed strains were generally susceptible to β-lactams; however, two human isolates were resistant to penicillin while one animal isolate was resistant to cefotaxime. Moreover, a substantial number of them were resistant to tetracycline, clindamycin, and erythromycin. Three strains showed induced-type MLSB multiresistance (one animal and two human isolates). Among the strains isolated from humans, three were resistant both to vancomycin and linezolid, whereas other three were resistant to tigecycline. Detailed antimicrobial susceptibility results are presented in Table 2.
Discussion
The first attempt to delineate the division criteria between S. dysgalactiae subspecies were undertaken by Vandamme et al. [22] in 1996. They were based on strain isolation source. According to this criterion, Lancefield C or G human isolates were classified to SDSE, whereas all strains isolated from animals were classified to SDSD. This classification reflected neither phenotypic nor genetic diversity of S. dysgalactiae species. Only 2 years later, in 1998, the division criteria were updated by Vieira et al. [23]. According to them, β-hemolytic Lancefield group C, G, or L strains were classified to SDSE, whereas nonhemolytic or α-hemolytic Lancefield group C strains were classified to SDSD. The latter division criteria have been used up to the present. However, many exceptions to these rules appear in literature, e.g., SDSE strains with Lancefield group A surface antigens [2, 20]. Moreover, not every SDSE strain is β-hemolytic on sheep blood agar [7]. Because of these ambiguities, some researchers continue to take into account the source of isolation as a division criterion. Therefore, animal infections are being assigned to SDSD strains, whereas human infections are being assigned to SDSE. Despite the fact that the etiological factor of the majority of human infections reported in literature is SDSE [17, 21] and of animal infections—SDSD [1, 10, 18], some cases contradicting this rule were reported, e.g., human infections caused by SDSD [14] and animal infections caused by SDSE [15].
The results of identification of the group of 36 isolates presented in this study question the subspecies division criteria described both by Vandamme et al. [22] and by Vieira et al. [23].
The conducted research suggests that the S. dysgalactiae hemolysis type on blood agar should not be longer considered a criterion in subspecies differentiation, at least in analyzed group of strains. Despite the fact that classification to Lancefield groups (C or G) confirms the SDSE characterization described by Vieira et al. [23], it is not an explicit and sufficient criterion. The same C-polysaccharide type (Lancefield group C) is also described as a feature of SDSD strains.
The presented results demonstrate that S. dysgalactiae strains isolated from clinical cases in animals might be identified as SDSE by means of currently available methods and their identification according to previously described criteria seems to be questionable.
Currently used S. dysgalactiae subspecies division criteria, described by Vieira et al., were established in the 1990s, so nearly 20 years ago. Therefore, currently available techniques allowing to analyze the relationship between strains much more precisely should be introduced. There is a possibility that in described period such changes in strains within S. dysgalactiae species occurred which obscure the differences between subspecies noted previously. Such a phenomenon is particularly apparent in bacteria which cross the animal-to-human interspecies barrier. The outline of evolutionary changes during this process was presented in 2007 in Nature by Wolfe et al. [25]. An adaptation to a new host is divided into several stages which sometimes might last unexpectedly short. One of the elements of this process, which has been already proven for other streptococci species [12], is horizontal gene transfer (HGT) from related species already adapted to human host. Horizontal gene transfer between S. pyogenes and S. dysgalactiae [8, 13] has already been recorded. It might lead to the emergence of a new, dangerous pathogen for human with the ability to be transferred via companion animals. We believe that presented results from relatively small group of strains should initiate the discussion about subspecies division criteria re-evaluation.
Antimicrobial susceptibility profiles of human and animal SDSE isolates presented in this study show that multiresistant strains emerge among human isolates. Some of them are also resistant to relatively new antibiotics which additionally enhance the threat.
Due to an increasing genetic variability and phenotypic variability of S. dysgalactiae strains isolated both from humans and animals as a result, the division into subspecies established many years ago seems to be questionable, especially when the division criteria are so underspecified. At this moment also, reflections about pan-genome and the general problem of bacterial species definition [11] might be taken into consideration. Although, if the division into two subspecies remains, which in the light of the presented data might be clinically negligible, the division into subspecies should base also on genetic analyses by means of DNA sequences unique for the subspecies (DNA fingerprint), including RISA method presented in this study as well as 16S rRNA genes sequencing. The latter method is also the basis for a phylogenetic relationship tree published in the latest issue of Bergey’s manual of systematic bacteriology [24].
References
Abdelsalam M, Fujino M, Eissa AE, Chen SC, Warda M (2015) Expression, genetic localization and phylogenic analysis of NAPlr in piscine Streptococcus dysgalactiae subsp. dysgalactiae isolates and their patterns of adherence. J Adv Res 6:747–755
Brandt CM, Haase G, Schnitzler N, Zbinden R, Lütticken R (1999) Characterization of blood culture isolates of Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield’s group A antigen. J Clin Microbiol 37:4194–4197
Brandt CM, Spellerberg B (2009) Human infections due to Streptococcus dysgalactiae Subsp. equisimilis. Emerg Infect 49:766–772
Caballero AR, Lottenberg R, Johnston KH (1999) Cloning, expression, sequence analysis and characterization of streptokinases secreted by porcine and equine isolates of Streptococcus equisimilis. Infect Immun 67:6478–6486
Cherkaoui A, Emonet S, Fernandez J, Schorderet D, Schrenzel J (2011) Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of beta-hemolytic streptococci. J Clin Microbiol 49:3004–3005
Clinical and Laboratory Standards Institute (2014) M100-S24: Performance standards for antimicrobial susceptibility testing
Dierksen KP, Tagg JR (2000) Haemolysin-deficient variants of Streptococcus pyogenes and S. dysgalactiae subsp. equisimilis may be overlooked as aetiological agents of pharyngitis. J Med Microbiol 49:811–816
Gherardi G, Imperi M, Palmieri C, Magi G, Facinelli B, Baldassarri L et al (2014) Genetic diversity and virulence properties of Streptococcus dysgalactiae subsp. equisimilis from different sources in Italy. J Med Microbiol 63:90–98
Hassan A, Khan IU, Lammler C (2003) Identification of Streptococcus dysgalactiae strains of Lancefield’s group C, G and L by polymerase chain reaction. J Vet Med Ser B 50:161–165
Jordal S, Glambek M, Oppegaard O, Kittang BR (2015) New tricks from an old cow: infective endocarditis caused by Streptococcus dysgalactiae subsp. dysgalactiae. J Clin Microbiol 53:731–734
Konstantinidis KT, Ramette A, Tiedje JM (2006) The bacterial species definition in the genomic era. Philos Trans R Soc B Biol Sci 361:1929–1940
Lun ZR, Wang QP, Chen XG, Li AX, Zhu XQ (2007) Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect Dis 7:201–209
McNeilly CL, McMillan DJ (2014) Horizontal gene transfer and recombination in Streptococcus dysgalactiae subsp. equisimilis. Front Microbiol 5:1–6
Park MJ, Eun IS, Jung CY, Ko YC, Kim YJ, Kim CK et al (2012) Streptococcus dysgalactiae subspecies dysgalactiae infection after total knee arthroplasty: a case report. Knee Surg Relat Res 24:120–123
Preziuso S, Laus F, Tejeda AR, Valente C, Cuteri V (2010) Detection of Streptococcus dysgalactiae subsp. equisimilis in equine nasopharyngeal swabs by PCR. J Vet Sci 11:67–72
Preziuso S, Pinho MD, Attili AR, Melo-Cristino J, Acke E, Midwinter AC et al (2014) PCR based differentiation between Streptococcus dysgalactiae subsp. equisimilis strains isolated from humans and horses. Comp Immunol Microbiol Infect Dis 37:169–172
Rantala S, Vahakuopus S, Vuopio-Varkila J, Vuento R, Syrjanen J (2010) Streptococcus dysgalactiae subsp. equisimilis bacteremia, Finland, 1995–2004. Emerg Infect Dis 16:843–846
Rato MG, Nerlich A, Bergmann R, Bexiga E, Nunes SF, Vilela CL et al (2011) Virulence gene pool detected in bovine group C Streptococcus dysgalactiae subsp. dysgalactiae isolates by use of a group A S. pyogenes virulence microarray. J Clin Microbiol 49:2470–2479
Suemori S, Sawada A, Komori S, Mochizuki K, Ohkusu K, Takemura H (2010) Case of endogenous endophthalmitis caused by Streptococcus equisimilis. Clin Ophthalmol 4:917–918
Tanaka D, Isobe J, Watahiki M, Nagai Y, Katsukawa C, Kawahara R et al (2008) Genetic features of clinical isolates of Streptococcus dysgalactiae subsp. equisimilis possessing Lancefield’s group A antigen. J Clin Microbiol 46:1526–1529
Tsai CT, Chi CY, Ho CM, Lin PC, Chou CH, Wang JH et al (2013) Correlation of virulence genes to clinical manifestations and outcome in patients with Streptococcus dysgalactiae subspecies equisimilis bacteremia. J Microbiol Immunol Infect 47:462–468
Vandamme P (1996) Taxonomic study of Lancefield streptococcal groups C, G and L. Int J Syst Bacteriol 46:774–781
Vieira VV, Teixeira LM, Zahner V, Momen H, Facklam RR, Steigerwalt AG et al (1998) Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int J Syst Bacteriol 48:1231–1243
Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F et al (2009) Bergey’s manual of systematic bacteriology—volume 3: the firmicutes. Springer, New York
Wolfe ND, Dunavan CP, Diamond J (2007) Origins of major human infectious diseases. Nature 447:279–283
Żabicka D, Izdebski R, Hryniewicz W (2009) Rekomendacje doboru testów do oznaczania wrażliwości bakterii na antybiotyki i chemioterapeutyki: Oznaczanie wrażliwości ziarniaków Gram-dodatnich z rodzaju Streptococcus spp., Krajowy Ośrodek Referencyjny ds. Lekowrażliwości Drobnoustrojów, Narodowy Instytut Leków, Warszawa
Acknowledgments
This study was funded by Medical University of Łódź (Grant No 502-03/3-012-03/502-34-051).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ciszewski, M., Zegarski, K. & Szewczyk, E.M. Streptococcus dysgalactiae subsp. equisimilis Isolated From Infections in Dogs and Humans: Are Current Subspecies Identification Criteria accurate?. Curr Microbiol 73, 684–688 (2016). https://doi.org/10.1007/s00284-016-1113-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1113-x