Abstract
Genome recoding with bias codons (synonymous rare codons) or codon pair bias is being used as a method to attenuate virulence mostly in viruses. The target gene chosen for attenuation in general in bacteria is mostly toxin or virulence gene. We have used RNA chaperone hfq, a global post-transcriptional regulator of bacterial gene expression that regulates about 20 % genes in Salmonella, as the target of recoding. The hfq gene was recoded by replacing the codons of hfq gene with synonymous rare codons. Recoding decreased the expression of Hfq protein about two-fold in the mutant as compared to the parent strain. Recoding did not affect growth kinetics, but in growth competition the mutant strain was outcompeted by the parent strain. There was significant decrease in survivability of mutant strain in macrophage as compared to the parent strain. The biofilm formation was significantly impaired in case of recoded mutant. The mutants were also less motile as compared to the parent strain. Intraperitoneal infection of mice with the mutant strain had shown better survival as compared to parent strain. The results show that recoding is an effective method of reducing virulence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The reduction in cost of gene synthesis has given an impetus to synthetic biology. New innovations are being tried, and one application where it has been successfully used is in making large-scale synonymous changes in the genome of viruses to attenuate pathogenicity [5, 7, 23–25]. Recoding genome of a pathogen by using synonymous codons is based on assumptions that codon usage of genes which are expressed in high and low amount in an organism is different [29]. The fact that many pathogens whose genome has been recoded have shown reduced virulence, testify above assumption [21, 23, 27, 29]. In codon bias (CB) which we have used in this study, rare synonymous codons replace all the codons of the target gene without changing the protein sequence. Codon bias was applied successfully to reduce pathogenicity of polio virus [5, 25]. Codon pair bias another variant of codon bias is based on differential use of adjacent codons and is independent of CB and has been similarly shown to reduce pathogenicity [21, 23, 27]. Both the approaches produce a protein which has identical amino acid sequence despite different nucleic acid sequences and thus an immune response similar to original pathogen [29].
Hfq has a diverse role in bacterial physiology and control of gene expression within bacterial cells [34]. Hfq acts as a global post-transcriptional regulator of gene expression [37]. About 20 % of genes of Salmonella are reported to be regulated by hfq [6]. The absence of RNA chaperone hfq deregulates more than 70 abundant proteins including the major outer membrane proteins [11, 33]. This protein is also involved in the expression and secretion of virulence factors in Salmonella Typhimurium [33]. Thus, hfq is a pleiotropic gene regulator and loss of hfq results in diverse phenotypes [6, 38] that compromise the fitness and virulence of many pathogenic bacteria.
Salmonella enterica serovar Typhimurium is a gram-negative, facultative anaerobic, intracellular pathogen of the family Enterobacteriaceae [30]. It is one of the most important food borne pathogens [13, 20] and a leading cause of gastroenteritis throughout the world. S. Typhimurium can cause disease in multiple hosts such as cattle, pigs, horses, sheep, poultry, and rodents including humans [30]. Contaminated animal products are major sources of human infection by Salmonella [2].
In this study, we have changed the codon usage pattern of hfq gene of Salmonella Typhimurium and show that recoding alters the phenotype and pathogenicity of recoded organism.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Growth Media
Salmonella Typhimurium PM 45 strain used in this study was of poultry origin and was provided by Dr. Mumtesh Saxena, College of Veterinary Science, GBPUAT, Pant Nagar, India. Plasmids used and generated in this study are listed in Table 1. Primers used in this study are listed in Table 2 and were designed from NCBI Reference Sequence: NC_003197.1. The bacteria strain E. Coli DH5α-λ pir used was gifted by Dr. Andrew Camilli, Tufts University. Boston, MA 02111-1817. pDS132 used in this study was a kind gift from Dominique Schneider, Université Joseph Fourier, France. When required, media were supplemented with antibiotics at the following concentrations: 100 µg ml−1 ampicillin, 30 µg ml−1 chloramphenicol, and 10 µg ml−1 chloramphenicol.
General Molecular Biology Methods
The plasmid DNA was isolated by using geneJet plasmid Miniprep kit (Fermentas, India) as per manufacturer’s instruction. Restriction digests and ligation reactions were carried out according to the manufacturer’s instructions using enzymes obtained from Fermentas. DNA was introduced into S. Typhimurium by electroporation using MicroPulser (Bio-Rad USA). PCR amplification of DNA was performed using High fidelity DNA polymerase (Roche) and, when required, PCR amplified products were gel purified using PureLink quick gel extraction kit (Invitrogen).
Design and Construction of Recoded hfq Gene of Salmonella Typhimurium
Codon usage table of Salmonella Typhimurium at http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=99287 was used for identifying rare codons used for protein synthesis in Salmonella Typhimurium (Table 1, supplementary materials). Table 1 gives the number of each codon and also the frequency of each codons per thousand codons. The codon of an amino acid that was least used in the genome was used as rare codons. Rare codons used are listed in table (Table 2, supplementary materials). Computational analysis was done on both original and recoded sequences on Vienna RNA website (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi and RNA promo (http://genie.weizmann.ac.il/pubs/rnamotifs08/rnamotifs08_predict.html). The recoded hfq gene and the un-recoded flanking regions (86 bp left flank, 186 bp right flank) were synthesized through vendor (Genescript). The sequence of recoded gene is given in Fig. 1.
Generation of Single Mutant Salmonella Typhimurium
The strategy adopted for generating homologous recombinant is shown in Fig. 2. The recoded hfq gene was cloned into pDS132 vector and electroporated into Salmonella Typhimurium. The single crossover was selected by chloramphenicol selection. Double crossover was selected on 10 % sucrose. The clones which were chloramphenicol sensitive and sucrose resistant were screened for the presence of recoded genes by PCR. The recoded gene from the mutant was amplified by PCR and sequenced through vendors to confirm orientation and the presence of recoded genes in the mutant.
The strategy used for generating homologous recombinant. 2.3 kb region of Salmonella Typhimurium (comprised 1 kb flanking regions covering 309 bp targeted region for recoding) cloned into pUC18 and the resultant plasmid was called pUChfq. Recoded region (309 bp) containing unrecoded flanking region with unique Sgr AI and Bsr GI sites in flanking region was custom made through vendors and was provided in pUC57 and was directionally cloned into Sgr AI and Bsr GI site of pUChfq. From the resultant plasmid (pUCmuthfq), the insert was removed with Sal I and was cloned into Sal I site of pDS132. The resultant plasmid (pDSmuthfq) was electroporated into Salmonella Typhimurium (STM), the chloramphenicol resistant clones (STM cam R) were next sucrose selected. From the resultant colonies, chloramphenicol-sensitive and sucrose-resistant (STM CamS SucR) colonies were picked up and screened for the presence of recoded gene by PCR
Antisera
Hyperimmune serum was produced in rabbits by subcutaneous inoculation with 100 μg of recombinant Hfq protein in Freund’s complete adjuvant (FCA). Subsequent inoculation was done on 14th, 21st, and 28th day. The presence of antibody in the serum was confirmed by Western blot with recombinant purified protein.
Western Blot Detection of Hfq
The Hfq protein was detected by Western blot in bacterial lysates of wild and mutant Salmonella Typhimurium as described previously [8]. The blot density was measured using Quantity one (Biorad).
Growth Kinetics and Growth Competition Assay
A single colony of S. Typhimurium inoculated into 5 ml of LB broth and kept overnight at 37 °C with constant shaking at 180 RPM. 50-ml broth was then inoculated with 50 μl of the overnight culture and was kept in a shaking incubator. OD 600 was measured at 1-h interval for 10 h against LB broth as blank. Growth competitions were carried out in LB by competing the wild and mutant strains as per the protocol described by Samhita et al. [31]. The numbers of mutant and parent strains were enumerated by colony PCR from random-picked colonies.
Macrophage Survival Assay
The protocol described by Sittka et al. [33] was used. The RAW 264.7 cell line was seeded in a 24-well plate at a concentration of 1 × 105 cells per well, 12 h prior to infection with the bacteria. The bacteria were grown in LB till stationary phase and then added to macrophages at a multiplicity of infection of 1. The plate was centrifuged at 250 g for 10 min (at 37 °C) and incubated at 37 °C for 20 min in an atmosphere containing 5 % CO2. After 30 min of infection, the cells were washed three times with PBS and RPMI containing 50 μg/ml of gentamicin was added to kill non-invasive bacterial cells followed by incubation for 1 h. Then the cells were washed with PBS and lysed with 0.1 % Triton X-100. The number of intracellular bacteria was determined by plating the different dilutions in HEA agar plate and expressed in per cent related to the input. Experiments were carried out in triplicates.
Biofilm Formation Assay
The biofilm-forming ability of the wild and mutant strains was assayed as per the protocol described by Ngwai et al. [26]. The assay was performed in a microtitre plate without shaking. Bacteria were grown in LB broth overnight at 37 °C. Then 10 µL of this overnight culture was inoculated into a flat-bottom 96-well polystyrene tissue culture plate containing 90 µL of LB broth followed by an incubation at 37 °C for 24 h. Cultures were then removed and rinsed 3 times with sterile distilled water to remove unattached bacteria. The wells were air dried at 37 °C, and adherent bacteria were stained at room temperature with 200 µL of 1 % aqueous solution of crystal violet for 20 min. The dye was removed from the wells and was rinsed 3 times with sterile distilled water followed by drying as before. Then 300 µL of dimethylsulfoxide was added to each well and the absorbance was measured at 600 nm. Results are means of three independent experiments.
Swimming Motility Assay
Swimming motility assay was performed as per the protocol described by Monteiro et al. [22]. The bacteria grown overnight at 37 °C on HEA plate were stab inoculated on 0.3 % LB without salt, agar plate using a toothpick. The plates were incubated at 28 °C for 7 h. The diameters of migrating bacteria from the point of inoculation (turbid zone) were measured. The results shown are representative of at least three independent experiments.
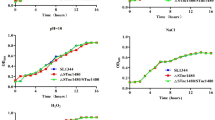
Stress Tolerance Assay
The stress tolerance assay was performed as per the protocol of Roscetto et al. [28]. Cultures were grown for 3 h at 37 °C in LB broth, harvested by centrifugation, washed once with phosphate-buffered saline (PBS). The culture was resuspended in water to an optical density at 600 nm (OD) of 1.0. Serial dilutions were pour plated onto HEA plates, supplemented with 5 % ethanol. Plates were incubated at 37 °C for 48 h. The results shown are representative of at least three independent experiments.
Survivability Study in Mice
Animal experimentation was carried out strictly as per the guidelines issued by Committee for the purpose of experiments on animals and was approved by institute animals ethics committee. The animals were observed twice daily and were euthanized by cervical dislocation after giving Phenobarbital sodium 200 mg/kg by i/p route. The terminal morbidity or moribund state was the endpoints for terminating animal experiment. Few animals died at night. The cause of death in all cases of mortality was established as Salmonellosis both by post-mortem examination and isolation of causative organism.
Six- to eight-week-old female BALB/c mice (18-20 g) obtained from the National Centre for Laboratory Animal Sciences (NCLAS), Hyderabad, India were used for studying the survival of mice upon infection with the mutant. The mice were kept for 10 days for acclimatization. All animal procedures were approved by Institute Animal Ethics Committee. The mice (14 in each group) were infected intraperitoneally (I/P) with 104 CFU of S. Typhimurium and the mice were observed twice daily for mortality and morbidity.
Statistics
The in vitro data were analyzed by Student’s t test. The survival curve analysis was done by Kaplan–Meier estimate. Results were determined to be statistically significant at a P value of less than 0.05.
Results
Computational Analysis of Recoded and Parent hfq Sequence
Most parameters that affect RNA degradation and translation efficiency primarily depend on UTR sequences [15] since we did not touch UTR sequence;, we did not foresee any effect on RNA degradation despite that we looked for motifs introduced by recoding (Table 3, supplementary materials). The recoding has not introduced any motifs which would affect degradation of the RNA. However, we did find substantial difference in the -∆G of optimal secondary structure acquired by the recoded sequence(−∆G = 43.90 kcal mol−1) and the original sequence (−∆G = 93.70 kcal mol−1) (Fig. 1, supplementary material). The secondary structure in the coding region has been shown to reduce the rate of translation [14, 16, 32] but there are reports showing that strong secondary structure is positively correlated with high protein level.
Recoding of hfq and Generation of Homologous Recombinant
The strategy adopted (Fig. 2) for generating homologous mutant resulted in many sucrose-resistant clones. From 22 sucrose-resistant colonies screened, only 5 were chloramphenicol-sensitive colonies and one of them was positive for the recoded hfq gene in PCR. The sequencing further confirmed the orientation and sequence of the recoded hfq.
Growth Characteristics
The in vitro growth kinetics did not reveal significant difference between mutant and parent strain (Fig. 3a). The experiment was repeated three times with very similar results. In growth competition assay between mutant and parent strain, a significant difference in their relative growth was observed (Fig. 3b). Competitive growth assay indicates relative fitness of the competing strains in nutrient depletion condition and the result suggests that mutant fitness to survive in nutrient depletion condition is compromised.
a In vitro growth kinetics. The results shown here are mean values of three independent experiments. b Growth competition between parent and Muthfq strain. Co-culturing equal inocula of parent and Muthfq mutant. The mean values of three independent experiments are shown along with standard error. c Western blot detection of Hfq protein in bacterial lysate. d Macrophage survival assay. Intracellular survival of parent and mutant strains in RAW 264.7 cell line. The results are means of three independent experiments along with standard error. e Biofilm-forming ability of parent and mutant strains. The results are means of three independent experiments along with standard error
Expression of Hfq in Mutant- and Parent-Type Salmonella Typhimurium
The Hfq protein expression in mutant and parent strain was assessed by Western blot (Fig. 3c). The Western blot had shown 1.78-fold reduction in the expression of Hfq protein in the mutant strain as compared to the parent strain.
Survival Inside Macrophages
The survival assay within macrophages was studied in murine macrophage cell line RAW264.7. The results indicated significant decrease in survival of mutant within macrophages (Fig. 3d). We did not do macrophage uptake assay so could not say if the difference is due to difference in uptake or the capacity to survive within macrophage; however, the decrease in number of bacteria in the macrophage indicates lower pathogenicity of the mutant strain compared to parent strain.
Biofilm-Forming Ability
The biofilm assay which again is related to virulence shows a significant decrease in the capacity of mutant to form biofilm as compared to parent strain (Fig. 3e). The result again shows a significant decrease in pathogenicity of mutant strain as compared to parent strain.
Swimming Motility, Stress Tolerance
The effect of recoding hfq on motility of Salmonella Typhimurium was checked on 0.3 % LB agar. The hfq mutant had significant decrease in the motility than the parent strain (Fig. 4a). The wild and the mutant were also exposed to stress by adding ethanol to the medium. However, no significant difference between stress tolerance of parent and mutant strain was observed (data not shown).
In Vivo Survival Study
The mice infected with parent strain died within 6 days. The mice infected with hfq mutant had shown better survival; 35.71 % mice survived by 6th day and 14.28 % mice survived the infection till the end of experiment on 21st day (Fig. 4b). The cause of death in all the mice was established as Salmonellosis by post-mortem examination. In all cases of death, Salmonella Typhimurium was isolated from the liver, spleen, and intestine of the mice that died during the study. No Salmonella were recovered from the healthy control.
Discussion
In this study, we have evaluated the effect of recoding transcriptional regulator hfq on pathogenicity of Salmonella Typhimurium. In an earlier study by Coleman et al. [8], they had shown that recoding toxin gene (pneumolysin) of Streptococcus pneumoniae reduces pathogenicity. In Pasteurella multocida recoding fis gene with synonymous rare codon had decreased pathogenicity [18].
Hfq regulates the expression of 20 % gene in Salmonella [6]. The hfq deletion mutant exhibits pleiotropic phenotypes such as decreased growth rate, reduced survival in stress conditions, and attenuation of virulence in a number of species [3, 9, 12, 19, 35]. The hfq deletion mutant is highly attenuated in mice and had shown protection at very high dose (108 cfu) in mice [1]. It has also been shown safe at this dose in pregnant mice. At lower dose it does not give adequate protection [1].
The effect of hfq deletion on growth rate of Salmonella strains has been marginal [28, 33]. We had also observed no difference in growth kinetics of mutant and parent strains. The effect of hfq deletion on growth competition has not been reported. We had observed a significant decrease in fitness of hfq mutant to compete against parent strain in nutrient depletion condition. This may also be true for hfq deletion mutant.
Survival within macrophages is an indicator of virulence [10]. The hfq knockout mutants of Salmonella Typhimurium or Y. pestis show reduced survival in macrophages [12, 33]. Hfq also affects biofilm formation and motility of many organisms [17, 22, 33, 35, 36, 39]. Our results also show a decline in the ability to form biofilm and decrease in the motility of mutant strain as compared to parent strain.
The in vitro results of different parameters show a decrease in all the parameters studied, except for stress-tolerance test. The hfq deletion decreases stress tolerance in Salmonella [33]. This was the only parameter which had not shown a decrease in hfq-recoded mutant; the reason could be any but one possible explanation could be that the decrease in the concentration of Hfq was not sufficient to alter stress tolerance. The decrease in the other parameters studied can be linked to lower Hfq concentration in the mutant (Fig. 3c). Hfq is a transcriptional regulator and regulates large number of genes. Its below normal concentration in recoded mutant could be the reason for these results.
The in vivo result of mice survivability shows a decrease in the pathogenicity of the mutant strain, in line with the in vitro results.
The hfq is a small size gene (309 bp). The probable impact of recoding on this gene could be small because recoding it requires only a small number of rare codons, and their impact on availability of tRNA may be not very significant. Further, decreasing the expression of hfq gene is possible by employing both codon pair bias and codon bias simultaneously. However, we cannot decrease the expression beyond a certain level by using these approaches. However, it is possible to decrease virulence by targeting multiple genes. We are working on that direction.
The recoding may introduce changes in the secondary structure, which may delete or insert motifs that may have major effect on RNA stability and translation. Stability of RNA is primarily dependent on UTR sequence and not on ORF [3, 4]; since we have not changed the sequence of UTRs, we do not expect an impact on RNA stability. The secondary structure in the coding sequences is known to reduce translation rate [14, 16, 32]. The minimum free energy of folding of the recoded and native sequence shows lower stability of recoded sequences as compared to original sequences, and therefore, logically it should be expressed in higher amount contrary to what has been observed in this study (Fig. 1, supplementary materials); however there are reports linking abundance of proteins to stable secondary structure [40]. The decrease we had observed in the expression of HFQ could be thus linked to the usage of rare codons. The objective of recoding is to reduce the expression of a key metabolite to a level where pathogen is attenuated to such an extent that recoded pathogen can be used as vaccine. Many strategies can be employed like introducing secondary structure and motifs for RNA degradation in the UTRs, using a combination of synonymous rare and non-rare codons to increase the secondary structure so that translation is decreased.
We feel that genome recoding can be exploited to customize pathogenicity to a level appropriate for the use as vaccine, and thus it provides a new avenue for vaccine development.
References
Allam US, Krishna MG, Lahiri A, Joy O, Chakravortty D (2011) Salmonella enterica serovar Typhimurium lacking hfq gene confers protective immunity against murine typhoid. PLoS One 6(2):e16667. doi:10.1371/journal.pone.0016667
Amavisit P, Browning GF, Lightfood D, Anderson CS (2001) Rapid PCR detection of Salmonella in horse faecal samples. Vet Microbiol 79:63–74
Bai G, Golubov A, Smith EA, McDonough KA (2010) The importance of the small RNA chaperone Hfq for growth of epidemic Yersinia pestis, but not Yersinia pseudotuberculosis, with implications for plague biology. J Bacteriol 192:4239–4245
Bernstein JA, Khodursky AB, Lin P-H, Lin-Chao S, Cohen SN (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA 99(15):9697–9702
Burns CC, Shaw J, Campagnolietal R (2006) Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol 80(7):3259–3272
Chao Y, Vogel J (2010) The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13:24–33
Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S (2008) Virus attenuation by genome-scale changes in codon pair bias. Science 320(5884):1784–1787
Coleman JR, Papamichail D, Yano M, del Mar García-Suárez M, Pirofski L (2011) Designed reduction of Streptococcus pneumoniae pathogenicity via synthetic changes in virulence factor codon-pair bias. J Infect Dis 203(9):1264–1273
Ding Y, Davis BM, Waldor MK (2004) Hfq is essential for Vibrio cholerae virulence and down regulates sigma expression. Mol Microbiol 53:345–354
Fields PI, Swanson RV, Haidaris CG, Heffron F (1986) Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci 83:5189–5193
Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L (2006) Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol Microbiol 62(3):838–852
Geng J, Song Y, Yang L et al (2009) Involvement of the posttranscriptional regulator Hfq in Yersinia pestis virulence. PLoS One 4:e6213
Gillespie BE, Mathew AG, Draughon FA, Jayarao BM, Oliver SP (2003) Detection of Salmonella enterica somatic groups C1 and E1 by PCR-enzyme-linked immunosorbent assay. J Food Prot 66:2367–2370
Guisez Y, Robbens J, Remaut E, Fiers W (1993) Folding of the MS2 coat protein in Escherichia coli is modulated by translational pauses resulting from mRNA secondary structure and codon usage: a hypothesis. J Theor Biol 162:243–252
Jacobson A (1996) Interrelationships of the pathways of m RNA decay and translation in eukaryotic cells. Annu Rev Biochem 65:693–739
Klionsky DJ, Skalnik DG, Simoni RD (1986) Differential translation of the genes encoding the proton-translocating ATPase of Escherichia coli. J Biol Chem 261:8096–8099
Kulesus RR, Diaz-Perez K, Slechta ES, Eto DS, Mulvey MA (2008) Impact of the RNAchaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect Immun 76:3019–3026
Kumar A (2015) A novel approach for designing, construction and evaluation of live mutant Pasteurella multocida serotype B:2 as a vaccine candidate. Dissertation, Indian Veterinary Research Institute
Lybecker MC, Abel CA, Feig AL, Samuels DS (2010) Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 78:622–635
Malorny B, Hoorfar J, Bunge C, Helmuth R (2003) Multicenter validation of the analytic accuracy of Salmonella PCR: toward an international standard. Appl Environ Microbiol 69:290–296
Martrus G, Nevot M, Andres C, Clotet B, Martinez MA (2013) Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirol 10:78
Monteiro C (2012) Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol 9(4):489–502
Mueller S, Coleman JR, Papamichail D et al (2010) Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol 28(7):723–726
Mueller S, Coleman JR, Wimmer E (2009) Putting synthesis into biology: a viral view of genetic engineering through de novo gene and genome synthesis. Chem Biol. doi:10.1016/j.chembiol.2009.03.002
Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E (2006) Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol 80(19):9687–9696
Ngwai YB, Adachi Y, Ogawa Y, Hara H (2006) Characterization of biofilm-forming abilities of antibiotic-resistant Salmonella Typhimurium DT104 on hydrophobic abiotic surfaces. J Microbiol Immunol Infect 39:278–291
Nouëna CL, Brocka LG, Luongoa C et al (2014) Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc Natl Acad Sci USA 111(36):13169–13174
Roscetto E, Angrisano T, Costa V et al (2012) Functional characterization of the RNA chaperone Hfq in the opportunistic human pathogen Stenotrophomonas maltophilia. J Bacteriol 194(21):5864–5874
Runco LM, Stauft CB, Coleman JR (2014) Tailoring the Immune response via customization of pathogen gene expression. J Pathog. doi:10.1155/2014/651568
Salyers A, Whitt D (2002) Bacterial pathogenesis: a molecular approach. ASM Press, Washington, pp 381–397
Samhita L, Nanjundiah V, Varshney U (2014) How many initiator tRNA genes does Escherichia coli need? J Bacteriol. doi:10.1128/JB.01620-14
Schmittgen TD, Danenberg KD, Horikoshi T, Lenz HJ, Danenberg PV (1994) Effect of 5-fluoro- and 5-bromouracil substitution on the translation of human thymidylate synthase mRNA. J Biol Chem 269:16269–16275
Sittka A, Pfeiffer V, Tedin K, Vogel J (2007) The RNA chaperone Hfq is essential for the virulence of Salmonella Typhimurium. Mol Microbiol 63:193–217
Sobrero P, Valverde C (2012) The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol 38(4):276–299
Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, Bläsi U (2003) Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog 35:217–228
Sousa SA, Ramos CG, Moreira LM, Leitão JH (2010) The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans. Microbiol 156:896–908
Valentin-Hansen P, Eriksen M, Udesen C (2004) The bacterial Sm-likeprotein Hfq: a key player in RNA transactions. Mol Microbiol 51:1525–1533
Vogel J (2009) A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol 71(1):1–11
Xiao-Gang W, Hui-Mei D, Tao T, Nan Y, Hong-You Z, Li-Quon Z (2010) Effect of the hfq gene on 2,4-diacetylphloroglucinol production and the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24. FEMS Microbiol Lett 309:16–24
Zur H, Tuller T (2012) Strong association between mRNA folding strength and protein abundance in S. cerevisiae. EMBO Rep 13:272–277
Acknowledgments
The authors acknowledge financial support received from Council for Scientific and Industrial Research (CSIR) and Indian Council of Agricultural research (ICAR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Behera, P., Kutty, V.H.M., Kumar, A. et al. Changing the Codon Usage of hfq Gene has Profound Effect on Phenotype and Pathogenicity of Salmonella Typhimurium. Curr Microbiol 72, 288–296 (2016). https://doi.org/10.1007/s00284-015-0951-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0951-2