Abstract
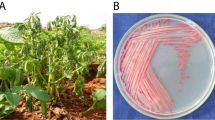
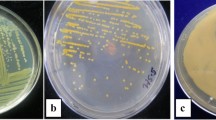
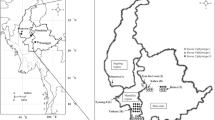
Bacterial wilt of tomato caused by Ralstonia solanacearum (Smith) Yabuuchi et al. (Microbiol Immunol 39:897–904, 1995) is a serious disease, which causes losses up to 60 % depending on environmental conditions, soil property, and cultivars. In present investigation, nucleotide sequences of virulence, hypersensitive response and pathogenicity (hrp) gene were used to design a pair of primer (Hrp_rs 2F: 5′-AGAGGTCGACGCGATACAGT-3′ and Hrp_rs 2R: 5′-CATGAGCAAGGACGAAGTCA-3′) for amplification of bacterial genome. The genomic DNA of 27 isolates of R. solanacearum race 1 biovar 3 & 4 was amplified at 323 bp. The specificity of primer was tested on 13 strains of R. solanacearum with other group of bacteria such as Xanthomonas oryzae pv. oryzae, X. campestris pv. campestris, and X. citri subsp. citri. Primer amplified DNA fragment of R. solanacearum at 323 bp. The sensitivity of the primer was 200 cfu/ml and improved further detection level by using non-specific enrichment medium casamino acids-pepton-glucose broth followed by PCR (BIO-PCR). Out of 130 samples of asymptomatic tomato plants, irrigation water, and soil collected from bacterial wilt infested field in different agro-climatic regions of India, R. solanacearum was detected from 86.9, 88.5, and 90.9 per cents samples using BIO-PCR, respectively. The primer was found specific for detecting viable and virulent strains of R. solanacearum and useful for the diagnosis of R. solanacearum in tomato seedlings and monitoring of pathogen in irrigation water and soil.




Similar content being viewed by others
References
Ayers SH, Rupp P Jr, Johnson WT (1919) A study of the alkali-forming bacteria in milk. U S Dep Agri Bull 782:1–139
Berg T, Tesoriero L, Hailstones DL (2005) PCR–based detection of Xanthomonas campestris pathovars in Brassica seed. Plant Pathol 54:416–427
Brumbley SM, Carney BF, Denny TP (1993) Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator. J Bacteriol 175:5477–5487
Elphinstone JG, Hennessy J, Wilson JK, Stead DE (1996) Sensitivity of different methods for the detection of Ralstonia solanacearum in potato tuber extracts. Bull OEPP/EPPO Bull 26:663–678
George MLC, Quinto V, Villamayor M, Nelson RJ (1996) A simple and rapid method for isolation of bacterial genomic DNA. Inter Rice Res Notes 21(2/3):84
Hayward AC (1994) Characteristics of Pseudomonas solanacearum. J Appl Bacteriol 27:27–265
Ito S, Ushijima Y, Fujii T, Tanaka S, Kameya-Iwaki M, Yoshiwara S, Kishi F (1998) Detection of viable cells of Ralstonia solanacearum in soil using a semi-selective medium and a PCR technique. J Phytopathol 146:379–384
Kang MJ, Lee MH, Shim JK, Seo ST, Shrestha R, Cho MS, Hain JH, Park DS (2007) PCR- based specific detection of Ralstonia solanacearum by amplified of cytochrome c1 signal peptide sequences. J Microbiol and Biotechnol 17(11):1765–1771
Lin CH, Hsu ST, Tzeng KC, Wang JF (2009) Detection of race 1 strains of Ralstonia solanacearum in field samples in Taiwan using a BIO-PCR method. Eur J Plant Pathol 124:75–85
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleic Acids Res 8:4321–4325
Pradhanang PM, Elphinstone JG, Fox RTV (2000) Sensitive detection of Ralstonia solanacearum in soil: a comparison of different detection techniques. Plant Pathol 49:414–422
Schaad NW, Jones JB, Chun W (2001) Laboratory guide for identification of plant pathogenic bacteria, 3rd edn. APS, St. Paul, p 245
Schonfeld J, Heuer H, Van Elsas JD, Smalla K (2003) Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of flic C fragments. Appl Environ Microbiol 69:7248–7256
Seal SE, Jackson LA, Young JPW, Daniels MJ (1993) Differentiation of Pseudomonas solanacearum, Pseudomonas syzygii, Pseudomonas pickettii and the blood disease bacterium by partial 16S rRNA sequencing: construction of oligonucleotide primers for sensitive detection by polymerase chain reaction. J Gen Microbiol 139:1587–1594
Singh D, Dhar Shri (2011) Bio-PCR based diagnosis of Xanthomonas campestris pathovars in black rot infected leaves of crucifers. Indian Phytopath 64(1):7–11
Singh D, Sinha S, Yadav DK, Sharma JP, Srivastava DK et al (2010) Characterization of biovar/races of Ralstonia solanacearum, the incitant of bacterial wilt in solanaceous crops. Indian Phytopath 63(3):261–265
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Bio Evol 28:2731–2739
Van der Wolf JM, Vriend SGC, Kastelen P, Nijhuis EH, Van Bekkum PJ, Van Vuurde JVL (2000) Immunofluorescence colony- staining (IFC) for detection and quantification of Ralstonia ( Pseudomonas) solanacearum biovar 2 (race 3) in soil and verification of positive results by PCR and dilution plating. Eur J Plant Pathol 106:123–133
Yabuuchi E, Kosako Y, Yano I, Hotta I, Nishiucliy Y (1995) Transfer of two Burkholderia and an Alcaligenes species to Ralstonia General Nov: proposal of Ralstonia pikettii (Ralston, palleroni and Doudoroff 1973) Comb. Nov, Ralstonia solanacearum (Smith1896) Comb. Nov. Microbiol Immunol 39:897–904
Acknowledgments
The authors are grateful to The Director, IISR, Calicut and Indian Council of Agricultural Research, New Delhi for providing financial support under outreach Project on “Phytophthora, Fusarium and Ralstonia Diseases of Horticultural and Field Crops” to conduct various experiments. The authors are also thankful to Dr. R. K. Jain, Head, Division of Plant Pathology, IARI New Delhi for his keen interest and help throughout the course of these investigation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, D., Sinha, S., Yadav, D.K. et al. Detection of Ralstonia solanacearum from Asymptomatic Tomato Plants, Irrigation Water, and Soil Through Non-selective Enrichment Medium with hrp Gene-Based Bio-PCR. Curr Microbiol 69, 127–134 (2014). https://doi.org/10.1007/s00284-014-0566-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0566-z