Abstract
Gene-nominated cj0183 was identified in Campylobacter jejuni NCTC 11168 and in two human isolates 81116 and 81-176. It encodes a protein which shows partial homology to TlyC of Brachyspira hyodysenteriae. The aim of this work was to determine the mechanisms of gene regulation by cloning DNA fragments lying upstream of the cj0183 gene. The β-galactosidase activity determined for the strain harboring the plasmid with the fragment upstream of cj0183 indicated the presence of a promoter in this DNA region. Mutations in cj0183 -10 region, -16 region, and -35 region resulted in changes in gene transcription.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Campylobacter jejuni is a major cause of bacterial diarrhea; yet, in comparison to other enteric pathogens still little is known about gene regulation in this bacteria. Only a few C. jejuni promoters have been experimentally confirmed so far [6, 7, 12, 17]. The C. jejuni operon structure is not well described. In silico analysis of the C. jejuni genome by Petersen et al. [10] revealed that the operon structure may be more expanded than previously described. The A+T content of the C. jejuni genome is very high (70 %) and genes from this bacterium are often difficult to clone and analyze in Escherichia coli [17]. Lack of expression in E. coli is mostly due to the absence of specific factors required for transcription. The consensus of the C. jejuni promoter sequence differs from the σ70 promoter consensus sequence of E. coli and other bacteria. It has been determined that the C. jejuni promoter contains three conserved regions: a -10 region (TATAAT) similar to E. coli, a -35 region (TTnAAGTnTT) completely different than in E. coli, and a -16 region (TTTTTTTG), which is typical for gram-positive bacteria and similar to the promoter found in E. coli promoters, named the extended -10 region [5, 9, 14, 17].
According to the suggestion of Wooldridge et al. [16], the product of the cj0183 gene found in the C. jejuni NCTC 11168 genome is implicated in colonization of epithelial cells of the chicken gut. Structure of the encoded protein contains domains characteristic for proteins with hemolytic activity. The cj0183 gene was found in all sequenced C. jejuni strains and all of the 69 C. jejuni and 16 Campylobacter coli PCR-tested strains isolated from chicken, dog, and pig stool samples [Sałamaszyńska-Guz, data not published]. The cj0183 gene was cloned to determine the role of the encoded protein in colonization of epithelial cells of the human gut. In vitro studies showed that mutation in the cj0183 gene does not influence adhesion to the Caco-2 cell line. Moreover, mutation in the cj0183 gene did not affect the hemolytic activity of the respective strains [11, 16]. Findings of Carvalho et al. [2] indicate that the TlyC protein from Leptospira (homolog of Cj0183) may contribute to binding to the extracellular matrix during host infection.
As little is known about the C. jejuni gene promoters, we decided to widen the knowledge concerning the function of specific C. jejuni promoter regions by analyzing the promoter of the Cj0183-encoding gene. In order to determine the regulation mechanism of this gene its upstream region was cloned in the pMW10 shuttle vector directly before the lacZ gene lacking its own promoter. The resulting recombinant plasmid was introduced into the C. jejuni 28C strain, in which the expression of the lacZ gene was studied. Consensus of the promoter sequence was determined after sequencing the cloned fragment that activated lacZ expression. The function of different motifs in the promoter region was also investigated.
Materials and Methods
Bacterial Strains, Plasmids, Media, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. The C. jejuni 28C strain was grown in microaerobic conditions at 42 °C on Mueller–Hinton (MH) agar (bioMérieux) containing 5 % (v/v) sheep blood. Strains were grown on media supplemented with kanamycin (Km; 25 μg/ml). E. coli strains were grown at 37 °C in LB broth or on LB agar (BIOCORP) supplemented with Km (25 μg/ml).
DNA Manipulations
Plasmid and chromosomal DNA extraction, restriction enzyme digestions, DNA ligation, agarose gel electrophoresis, and transformations were performed by standard procedures using enzymes supplied by Fermentas. Isolation of DNA from agarose gel was performed according to the manufacturer’s instructions (A&A Biotechnology).
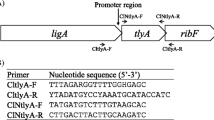
Oligonucleotide primers which were synthesized by Genomed S.A. Primer sequences are given in Table 2. DNA sequencing was performed by Genomed S.A.
Reporter Gene Assays
The C. jejuni promoter probe vector pMW10 contains a promoterless lacZ gene, and was previously used to quantify promoter activity in C. jejuni [17]. An overlapping fragment containing the cj0183 start codon and part of the upstream region was amplified by PCR using specific primers (Table 2) and C. jejuni 81-176 genomic DNA as template. The amplified PCR product was cloned into pMW10 using BamHI and BglII sites. The resulting plasmid was named p0183 (WT). Constructs were transformed into E. coli DH5α and C. jejuni 28C strains.
Site-Directed Mutagenesis
Standard-purified mutagenesis primers were designed following the QuickChange™ II XL site-directed mutagenesis kit guidelines (Stratagene). Mutant Δ -35 was constructed using primers M1 and M35; mutant Δ -35-16—primers M1 and M35-16; mutant M35-16-10—primers M1 and M35-16-10; mutant Subs/restr -10—primers M35-16-10 and Mrest-10; mutant Subs/restr -16—primers M35-16-10 and Mrest-16; mutant Subs/cons -16—primers M35-16-10 and Mcons-16; mutant Subs/cons -35—primers M1 and Mcons-35. Primer sequences are presented in Table 2. PCR products of the mutagenesis reactions were digested with DpnI and purified using the DNA Clean-Up kit (A&A Biotechnology). Subsequently, products were phosphorylated, ligated and used for electroporation of E. coli DH5α and C. jejuni 28C.
Transformation of Campylobacter jejuni
Campylobacter jejuni electrocompetent cells prepared as described by Wassenaar et al. [15] were mixed with plasmid DNA (0.5–5 μg). Electroporation was performed in Electro Cell Manipulator ECM 600 by applying 50 μF, 126 Ω, and 1.25 kV. Transformants were grown under microaerobic conditions at 42 °C on MH plates for 5 h and then plated on BHI agar plates supplemented with 5 % sheep blood and a selective antibiotic—Km. Transformants were grown for 2–5 days.
β-Galactosidase Assay
β-Galactosidase activity in E. coli and C. jejuni was measured by conversion of o-nitrophenyl-β-d-galactopyranoside into nitrophenol as described by Wösten et al. [17]. ß-Galactosidase activities were expressed in Miller units based on three independent experiments.
Primer Extension
Total RNA of wild-type C. jejuni strain 81-176 was isolated using the RNA protect RNeasy Mini kit (Qiagen) followed by DNase treatment. cDNA synthesis was performed using Fermentas RevertAid First Strand cDNA Synthesis Kit according to the manufacturer’s instruction. RT-PCRs were performed using the 183-6FAM primer (Table 2) and products were analyzed using Peak Scanner™ Software v 2.0 Applied Biosystems.
Sequence Data Analysis
Each construct which was sequenced by Genomed S.A. Amino acid sequences were aligned using Clustal W. Computational search of wild genome promoters was performed using RSAT (http://rsat.uib.ac.be/rsat) [3, 13].
Results and Discussion
Determination of β-Galactosidase Activity
The β-galactosidase activity assay was performed in E. coli DH5α and C. jejuni 28C cells, harboring the pMW10 recombinant plasmid, p0183 (WT), carrying an insert with the probable promoter region of the C. jejuni 81-176 cj0183 gene amplified using 0183P1 and 0183P2 primers. E. coli DH5α and C. jejuni 28C strains with the pMW10 plasmid alone served as negative controls.
In accordance with earlier reports [17], a significant variation of β-galactosidase activity between E. coli and C. jejuni, due to differences in their promoter sequences, was observed (P < 0.05). Activity of the enzyme was considerably lower in E. coli cells harboring the p0183 (WT) recombinant plasmid (39.7 ± 12.1) compared to the activity detected in C. jejuni (619.1 ± 93.7). Findings comply with literature data and result from differences in the genome G+C content of these bacteria, which reflects also dissimilarities in the structure of promoter regions [17]. Based on the β-galactosidase activity assay in E. coli and C. jejuni strains harboring the constructs, it seems that the upstream DNA sequence of the cj0183 gene most probably contains a promoter sequence.
Analysis of Nucleotide Sequences Upstream of the cj0183 Gene
The coding and promoter sequences of the cj0183 gene are highly conserved among sequenced C. jejuni genomes (99–100 % for coding and 100 % for promoter sequence). Based on bioinformatic analysis (RSAT) it seems highly probable that cj0183 remains under the control of the σ70-recognized promoter [13]. Sequence of the upstream region of cj0183 showed no homology to sequences recognized by other sigma factors of C. jejuni. The cj0183 promoter contains three typical conserved promoter motifs: -10, -16, and -35 [17], at an appropriate distance upstream from the transcriptional start site (Fig. 1). Position of the first T in the -10 motif of the predicted promoter in relation to the start codon is 42 bp; majority of -10 motifs predicted by Petersen et al. [10] start 30–43 bp upstream of the start codon.
We constructed various deletions mutants within the assumed promoter region by removing: (i) the whole promoter region (Δ-35-16-10), (ii) -16 and -35 motifs (Δ-35-16), or (iii) only the -35 motif (Δ-35) (Fig. 2). All of these deletions significantly affected the activity of the β-galactosidase reporter gene (Fig. 3). Deletion of the -35-16-10 and -35-16 region abolished completely and deletion of the -35 region decreased threefold β-galactosidase activity, indicating that these regions are involved in gene transcription under standard laboratory conditions.
Mutational analysis of the cj0183 promoter region. Quantitative analysis of lacZ expression of the cj0183 promoter deletion/substitution vector series in C. jejuni, including the construct carrying the wild-type cj0183 promoter (WT) as a positive control and the empty vector (control) as a negative control. Reactions were performed in triplicate, and standard deviations are marked by error bars. *P > 0.05
Genome Analysis of Putative Campylobacter jejuni Promoters and Experimental Validation of cj0183 Promoter Motifs
We used RSAT to perform genome analysis of putative C. jejuni 81-176 strain promoters under the following criteria: (i) both strands of DNA were scanned, (ii) TnAAGTnnn and TATAAT were the conserved -35 and -10 motifs, respectively, (iii) only hits located in noncoding sequences were counted, (iv) spacers between the -10 and -35 motifs in C. jejuni contained 15–19 bp [3, 13, 17]. We predicted promoter regions upstream of 146 reading frames (from 100 bp upstream to 1 bp downstream of the annotated start codon). Among them, 133 promoters were located on the chromosome, 10 on the Vir plasmid and 3 on the Tet plasmid. Our results indicate that promoters with a 18 bp (n = 55) spacer are dominant, which is in contrast to the study of Wösten et al. [17] reporting that promoters with a 17 bp spacer were the most prevailing. In our study, we determined that the promoter of the cj0183 gene has a 19 bp spacer (Table 3).
The importance of putative cj0183 promoter motifs was experimentally investigated by generating substitutions and deletions in these regions (Fig. 2), and performing subsequent transcriptional fusions of the mutated promoters with a promoterless lacZ reporter gene in pMW10 [17]. Effects of mutations made in the promoter region were then quantitatively assessed based on the activity of the β-galactosidase enzyme produced.
The C. jejuni 81-176 cj0183 promoter contains an optimal -10 region (TATAAT) compared to the consensus E. coli and C. jejuni sequence (TATAAT). Substitution within this motif by an unrelated sequence GGATCC rich in GC (Subs/restr -10) abolished β-galactosidase activity completely (Fig. 3). The study by Petersen et al. [10] indicated that the -10 motif is a highly conserved; thus, it is not surprising that substitution within this region seriously influences the activity of the promoter.
The predicted -35 motif (TTCAAATnTT) does not match the E. coli consensus sequence, but is similar to the predicted canonical C. jejuni -35 motif TTTAAGTnTT [17]. Alteration of the cj0183 -35 motif to the consensus sequence in the Subs/cons -35 strain increases β-galactosidase activity approximately twofold (Fig. 3). In contrast, as mentioned above, deletion of the -35 region decreases threefold β-galactosidase activity. Our studies determined that the -35 region is not essential for promoter activity; its removal significantly reduces, but does not abolish the activity of the examined promoter sequence. However, the promoter activity increased significantly above 1,000 Miller units, when the -35 region was constituted by the consensus sequence proposed by Wösten et al. [17]. Data published by Petersen et al. [10] describing an in silico analysis of the predicted promoter sequences of C. jejuni and H. pylori show a weakly conserved -35 motif. Despite that in C. jejuni the -35 region is not highly conserved, its strong compatibility with the consensus sequence increases promoter strength.
The spacer between -35 (TTCAAATnTT) and -10 (TATAAT) motifs in the cj0183 promoter counts 19 bp. Within this region we have found a -16 motif. The C. jejuni 81-176 cj0183 promoter contains a suboptimal -16 region (TTAATTGT) compared to the consensus C. jejuni sequence (TTTTTTTG). Alteration of the cj0183 -16 motif to the consensus sequence in the Subs/cons -16 mutant does not significantly affect β-galactosidase activity (Fig. 3). Our results allowed to establish that the -16 motif is important for transcription of the cj0183 gene. Mutation of the -16 motif to a AGATCTT sequence in Subs/restr -16 abolished β-galactosidase activity completely (Fig. 3).
The importance of the -16 TGn (TTTTTT TG) motifs in transcription initiation has been well documented in other bacteria [1, 4, 14]. The extended -10 TGn motif seems to play an important role in C. jejuni transcription since a third of predicted by Wösten et al. and about 13 % of computationally discovered (RSAT) C. jejuni promoters have this motif (Tables 3, 4). The TGn motif is found to compensate poor -10 hexamer and -35 element in E. coli [5, 9]. Detailed analysis of predicted C. jejuni promoters indicates a weaker -35 motif in promoters with TGn comparison with non-TG promoters. Furthermore, the -10 region resembles more the consensus of TG promoters (Table 4). This confirms the observation of Petersen et al. [10] that C. jejuni promoters contain a strongly conserved -10 region, but do not possess a conserved -35 region. Moreover, C. jejuni promoters have a strong periodic signal upstream of the -10 region that has not been reported before in bacterial promoters. It contains thymine bases every 10–11 bp and may play a role in environmental regulation of the gene expression level.
The -16 region of the cj0183 gene has two TG motifs—TAATTG TGTATAAT immediately upstream of the -10 hexamer at -13/-14 positions and TAAT TGTGTATAAT with a 2 nucleotide spacer (TGnn) at -15/-16 positions. The promoter in the Subs/cons -16 mutant (consensus -16 motif sequence) has a single nucleotide spacer between the -16 and -10 region (TGn) at -14/-15 positions and the same activity as the wild-type cj0183 gene promoter (Figs. 2, 3). The cj0183 promoter contains a G at the -13 position. Djordjevic [5] suggested significant conservation of this nucleotide in E. coli, but its importance has not been reported.
Further study may provide insight into the process of gene transcription in C. jejuni and may be relevant in understanding the molecular responses required to survive in the host. Therefore, to confirm the exact role of the -16 region and TG motif in C. jejuni gene transcription, single-base pair mutations in these regions will be required.
References
Burr T, Mitchel J, Kolb A, Minchin S, Busby S (2000) DNA sequence elements located immediately upstream of the -10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res 28:1864–1870
Carvalho E, Barbosa AS, Gómez RM, Cianciarullo AM, Hauk P, Abreu PA, Fiorini LC, Oliveira ML, Romero EC, Gonçales AP, Morais ZM, Vasconcellos SA, Ho PL (2009) Leptospiral TlyC is an extracellular matrix-binding protein and does not present hemolysin activity. FEBS Lett 583:1381–1385
Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED (2007) Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J Bacteriol 189:5108–5118
Davies BJ, de Vries N, Rijpkema SG, van Vliet AHM, Penn CW (2002) Transcriptional and mutational analysis of the Helicobacter pylori urease promoter. FEMS Microbiol Lett 213:27–32
Djordjevic M (2011) Redefining Escherichia coli σ70 promoter elements: -15 motif as a complement of the -10 motif. J Bacteriol 193:6305–6314
Jeon B, Itoh K, Ryu S (2005) Promoter analysis of cytolethal distending toxin genes (cdtA, B, and C) and effect of a luxS mutation on CDT production in Campylobacter jejuni. Microbiol Immunol 49:599–603
Kim M, Hwang S, Ryu S, Jeon B (2011) Regulation of perR expression by iron and PerR in Campylobacter jejuni. J Bacteriol 193:6171–6178
Korlath JA, Osterholm MT, Judy LA et al (1985) A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis 152:592–596
Mitchell JE, Zheng D, Busby SJW, Minchin SD (2003) Identification and analysis of ‘extended -10’ promoters in Escherichia coli. Nucleic Acid Res 31:4689–4695
Petersen L, Larsen TS, Ussery DW, On SL, Krogh A (2003) RpoD promoters in Campylobacter jejuni exhibit a strong periodic signal instead of a -35 box. J Mol Biol 326:1361–1372
Sałamaszyńska-Guz A, Klimuszko D (2008) Functional analysis of the Campylobacter jejuni cj0183 and cj0588 genes. Curr Microbiol 56:592–596
Thies FL, Weishaupt A, Karch H, Hartung HP, Giegerich G (1999) Cloning, sequencing and molecular analysis of the Campylobacter jejuni groESL bicistronic operon. Microbiology 145:89–98
van Helden J (2003) Regulatory sequence analysis tools. Nucleic Acids Res 31:3593–3596
Voskuil MI, Chambliss GH (1998) The -16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res 26:3584–3590
Wassenaar TM, Fry BN, van der Zeijst BA (1993) Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 30:131–135
Wooldridge KG, Jones M, Morris S, Carrasco M, Powel H, Forsythe S, Ala’Aldeen D (2004) Two-toxins related genes of C. jejuni have a role in colonisation of the chickens. CHRO: 78
Wösten MM, Boeve M, Koot MG, van Nuenen AC, van der Zeijst BA (1998) Identification of Campylobacter jejuni promoter sequences. J Bacteriol 180:594–599
Acknowledgments
Financial support was received from 5041002340011 (Warsaw University of Life Sciences—SGGW).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Salamasznska-Guz, A., Grodzik, M. & Klimuszko, D. Mutational Analysis of cj0183 Campylobacter jejuni Promoter. Curr Microbiol 67, 696–702 (2013). https://doi.org/10.1007/s00284-013-0420-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0420-8