Abstract
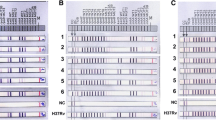
Mycobacterium tuberculosis has developed resistance to anti-tuberculosis first-line drugs. Multidrug-resistant strains complicate the control of tuberculosis and have converted it into a worldwide public health problem. Mutational studies of target genes have tried to envisage the resistance in clinical isolates; however, detection of these mutations in some cases is not sufficient to identify drug resistance, suggesting that other mechanisms are involved. Therefore, the identification of new markers of susceptibility or resistance to first-line drugs could contribute (1) to specifically diagnose the type of M. tuberculosis strain and prescribe an appropriate therapy, and (2) to elucidate the mechanisms of resistance in multidrug-resistant strains. In order to identify specific genes related to resistance in M. tuberculosis, we compared the gene expression profiles between the pansensitive H37Rv strain and a clinical CIBIN:UMF:15:99 multidrug-resistant isolate using microarray analysis. Quantitative real-time PCR confirmed that in the clinical multidrug-resistant isolate, the esxG, esxH, rpsA, esxI, and rpmI genes were upregulated, while the lipF, groES, and narG genes were downregulated. The modified genes could be involved in the mechanisms of resistance to first-line drugs in M. tuberculosis and could contribute to increased efficiency in molecular diagnosis approaches of infections with drug-resistant strains.



Similar content being viewed by others
References
Ali A, Hasan R, Jabeen K, Jabeen N, Qadeer E, Hasan Z (2011) Characterization of mutations conferring extensive drug resistance to Mycobacterium tuberculosis isolates in Pakistan. Antimicrob Agents Chemother 55(12):5654–5659
Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, Young DB (2011) Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog 7(11):e1002342
Bailey SL, Roper MH, Huayta M, Trejos N, Lopez Alarcon V, Moore DA (2011) Missed opportunities for tuberculosis diagnosis. Int J Tuberc Lung Dis 15(2):205–210, i
Betts JC, McLaren A, Lennon MG, Kelly FM, Lukey PT, Blakemore SJ, Duncan K (2003) Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob Agents Chemother 47(9):2903–2913
Castorena-Torres F, Bermudez de Leon M, Cisneros B, Zapata-Perez O, Salinas JE, Albores A (2008) Changes in gene expression induced by polycyclic aromatic hydrocarbons in the human cell lines HepG2 and A549. Toxicol In Vitro 22(2):411–421
Chaoui I, Sabouni R, Kourout M, Jordaan AM, Lahlou O, Elouad R, Akrim M, Victor TC, El Mzibri M (2009) Analysis of isoniazid, streptomycin and ethambutol resistance in Mycobacterium tuberculosis isolates from Morocco. J Infect Dev Ctries 3(4):278–284
Fritz C, Maass S, Kreft A, Bange FC (2002) Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect Immun 70(1):286–291
Fu LM, Fu-Liu CS (2007) The gene expression data of Mycobacterium tuberculosis based on Affymetrix gene chips provide insight into regulatory and hypothetical genes. BMC Microbiol 7:37
Fu LM, Shinnick TM (2007) Genome-wide analysis of intergenic regions of Mycobacterium tuberculosis H37Rv using Affymetrix GeneChips. EURASIP J Bioinform Syst Biol: 23054
Gagneux S (2009) Fitness cost of drug resistance in Mycobacterium tuberculosis. Clin Microbiol Infect 15(Suppl 1):66–68
Gao Q, Kripke KE, Saldanha AJ, Yan W, Holmes S, Small PM (2005) Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 151(Pt 1):5–14
Goh KS, Rastogi N, Berchel M, Huard RC, Sola C (2005) Molecular evolutionary history of tubercle bacilli assessed by study of the polymorphic nucleotide within the nitrate reductase (narGHJI) operon promoter. J Clin Microbiol 43(8):4010–4014
Guo JH, Xiang WL, Zhao QR, Luo T, Huang M, Zhang J, Zhao J, Yang ZR, Sun Q (2008) Molecular characterization of drug-resistant Mycobacterium tuberculosis isolates from Sichuan Province in china. Jpn J Infect Dis 61(4):264–268
Hillemann D, Rusch-Gerdes S, Richter E (2009) Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 47(6):1767–1772
Homolka S, Niemann S, Russell DG, Rohde KH (2010) Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog 6(7):e1000988
Ilghari D, Lightbody KL, Veverka V, Waters LC, Muskett FW, Renshaw PS, Carr MD (2011) Solution structure of the Mycobacterium tuberculosis EsxG.EsxH complex: functional implications and comparisons with other M. tuberculosis Esx family complexes. J Biol Chem 286(34):29993–30002
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31(4):e15
Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J (1997) Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35(4):907–914
Lee JM, Zhang S, Saha S, Santa Anna S, Jiang C, Perkins J (2001) RNA expression analysis using an antisense Bacillus subtilis genome array. J Bacteriol 183(24):7371–7380
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408
Maciag A, Piazza A, Riccardi G, Milano A (2009) Transcriptional analysis of ESAT-6 cluster 3 in Mycobacterium smegmatis. BMC Microbiol 9:48
Molina-Salinas GM, Ramos-Guerra MC, Vargas-Villarreal J, Mata-Cardenas BD, Becerril-Montes P, Said-Fernandez S (2006) Bactericidal activity of organic extracts from Flourensia cernua DC against strains of Mycobacterium tuberculosis. Arch Med Res 37(1):45–49
Park WD, Stegall MD (2007) A meta-analysis of kidney microarray datasets: investigation of cytokine gene detection and correlation with rt-PCR and detection thresholds. BMC Genomics 8:88
Peñuelas-Urquides K, Villarreal-Treviño L, Silva-Ramírez B, Rivadeneyra-Espinoza L, Said-Fernández S, Bermúdez de León M (2013) Measuring of Mycobacterium tuberculosis growth. A correlation of the optical measurements with colony forming units. Brazilian J Microbiol (in press)
Qamra R, Mande SC, Coates AR, Henderson B (2005) The unusual chaperonins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 85(5–6):385–394
Saviola B, Woolwine SC, Bishai WR (2003) Isolation of acid-inducible genes of Mycobacterium tuberculosis with the use of recombinase-based in vivo expression technology. Infect Immun 71(3):1379–1388
Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE 3rd, Wang H, Zhang W, Zhang Y (2011) Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333(6049):1630–1632
Sidders B, Withers M, Kendall SL, Bacon J, Waddell SJ, Hinds J, Golby P, Movahedzadeh F, Cox RA, Frita R, Ten Bokum AM, Wernisch L, Stoker NG (2007) Quantification of global transcription patterns in prokaryotes using spotted microarrays. Genome Biol 8(12):R265
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3
Somoskovi A, Parsons LM, Salfinger M (2001) The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res 2(3):164–168
Su T, Waxman DJ (2004) Impact of dimethyl sulfoxide on expression of nuclear receptors and drug-inducible cytochromes P450 in primary rat hepatocytes. Arch Biochem Biophys 424(2):226–234
Subbian S, Tsenova L, O’Brien P, Yang G, Koo MS, Peixoto B, Fallows D, Dartois V, Muller G, Kaplan G (2011) Phosphodiesterase-4 inhibition alters gene expression and improves isoniazid-mediated clearance of Mycobacterium tuberculosis in rabbit lungs. PLoS Pathog 7(9):e1002262
Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM, Jacobs WR Jr (1997) The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3(5):567–570
van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM et al (1993) Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31(2):406–409
Waddell SJ, Stabler RA, Laing K, Kremer L, Reynolds RC, Besra GS (2004) The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tuberculosis (Edinb) 84(3–4):263–274
WHO (2012) Global Tuberculosis Report 2012. World Health Organization, Geneva
Acknowledgments
This study was supported by the National Council for Science and Technology (CONACyT), Mexico, Grant No. 99792.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Peñuelas-Urquides, K., González-Escalante, L., Villarreal-Treviño, L. et al. Comparison of Gene Expression Profiles Between Pansensitive and Multidrug-Resistant Strains of Mycobacterium tuberculosis . Curr Microbiol 67, 362–371 (2013). https://doi.org/10.1007/s00284-013-0376-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0376-8