Abstract
In the last few years, numerous cases of multidrug-resistant Achromobacter xylosoxidans infections have been documented in immunocompromised and cystic fibrosis patients. To gain insights into the molecular mechanisms and mobile elements related to multidrug resistance in this bacterium, we studied 24 non-epidemiological A. xylosoxidans clinical isolates from Argentina. Specific primers for plasmids, transposons, insertion sequences, bla ampC, intI1, and intI2 genes were used in PCR reactions. The obtained results showed the presence of wide host range IncP plasmids in ten isolates and a high dispersion of class 1 integrons (n = 10) and class 2 integrons (n = 3). Four arrays in the variable region (vr) of class 1 integrons were identified carrying different gene cassettes as the aminoglycoside resistance aac(6′)-Ib and aadA1, the trimethoprim resistance dfrA1 and dfrA16, and the β-lactamase bla OXA-2. In only one of the class 2 integrons, a vr was amplified that includes sat2-aadA1. The bla ampC gene was found in all isolates, confirming its ubiquitous nature. Our results show that A. xylosoxidans clinical isolates contain a rich variety of genetic elements commonly associated with resistance genes and their dissemination. This supports the hypothesis that A. xylosoxidans is becoming a reservoir of horizontal genetic transfer elements commonly involved in spreading antibiotic resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Achromobacter spp. is a rarely nosocomial and community pathogen, being Achromobacter xylosoxidans the most frequent species among Achromobacter spp. isolates [6, 8, 18]. Many reports of A. xylosoxidans infections are documented in immunocompromised and cystic fibrosis (CF) patients, where its pathogenic role has not yet been properly clarified [7, 8]. In Argentina, the relative frequency of A. xylosoxidans among the uncommon non-glucose-fermenting gram-negative bacilli infections has been increasing reaching 66 % of total non-glucose-fermenting gram-negative bacilli infection isolates [18].
Although clinical A. xylosoxidans isolates usually show multiple drug resistance, the relative low attention paid to this pathogen resulted in poor understanding of their resistance mechanisms. Little is known about molecular mechanisms and transferable elements contributing to the acquisition and dissemination of antibiotic resistance determinants in A. xylosoxidans clinical isolates.
The aim of this study was to explore the occurrence of mobile elements related to antibiotic-resistance determinants among a collection of 24 non-epidemiological-related clinical isolates of A. xylosoxidans recovered in Argentina from six centers.
Materials and Methods
Bacterial Strains
Twenty-four non-epidemiological-related clinical isolates of A. xylosoxidans recovered in Argentina from six centers were used (Table 1). All isolates were identified using standard biochemical tests and API 20NE (Biomeriux), and the species level was confirmed by sequencing the 16S rRNA gene [19]. Clonal relationships analysis, using the macrorestriction technique, showed the presence of 15 different clones among the isolates included in the study (data not shown). The antibiotic susceptibility was performed by agar dilution method following the general recommendations of the Clinical and Laboratory Standards Institute (CLSI) [4].
DNA Techniques
Total DNAs were prepared and used as template for PCR reactions. PCR reactions were carried out using the GoTaq enzyme according to manufacturer’s instructions (Promega, Madison, WI), and the products were detected by agarose gel electrophoresis. To reveal the presence of transferable determinants associated to horizontal gene transfer, specific primers for plasmids (IncP, IncW, IncA/C, IncN, IncFII, repAci1), transposons (Tn1331, Tn3, Tn7), insertion sequences (IS) (IS26, IS440), and the bla ampC, intI1, and intI2 genes were used (Table 2). The selection of the mobile elements was based on its association with antibiotic-resistance determinants and also its distribution in our hospitals [12, 13, 16].
DNA Sequencing
PCR products were sequenced after purifying the DNA by using the Wizard SV Gel and PCR clean-up System kit according to the manufacturer’s directions (Promega, USA). Sequencing was performed on both DNA strands, using an ABIPrism 3100 BioAnalyzer equipment. The nucleotide sequences were analyzed using the Blast V2.0 software (http://www.ncbi.nlm.nih.gov/BLAST/).
Results and Discussion
The 24 A. xylosoxidans isolates studied exhibited the typical multiresistance profile previously described for this species, being the third and fourth-generation cephalosporins, fluoroquinolones, and aminoglycosides not active against Achromobacter spp. [18]. All isolates were susceptible to tazobactam, imipenem, and meropenem (Table S1 in Supplementary material).
Among the PCR reactions performed for the selected transferable elements, positive results were obtained in ten isolates (42 %) for the IncP plasmids, a wide host range and self-transmissible plasmid important in the dissemination of resistant genes around the world [11] (Table 1). Negative results were obtained for the other Inc groups searched (IncW, IncA/C, IncN, IncFII). Sequence analysis of the amplification products showed 99 % of identity in 200-bp length with the replication gene trfA (AN GU186864). The GC% of the trfA replication gene of IncP plasmid is 60.5 %, which is very similar to the GC% (67 %) of A. xylosoxidans. We also noticed in this study that most isolates containing IncP plasmids corresponded to nosocomial isolates (n = 9). In only one CF patient isolate (Ax72), an IncP plasmid was identified.
Regarding IS and transposons, positive results were obtained for IS26 (n = 2) and IS440 (n = 7) (Table 1), two ISs frequently associated to antimicrobial resistance genes and to classes 1 and 2 integrons [1, 2, 10], obtaining negative results for the transposons Tn1331, Tn3, and Tn7.
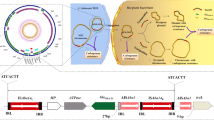
In addition, a high dispersion of class 1 integrons was found (42 %). Most of the positive isolates corresponded to nosocomial patient samples (n = 9), being only one positive isolate from a CF patient sample (Ax72). To characterize the vr of class 1 integrons, PCR cartography was carried out as previously described [12]. Four vr were identified, being all the arrays different to the previous arrays reported in this species (Table 1; Fig. 1). Among the gene cassettes identified in the class 1 integron context, aminoglycosides-resistance genes aac(6′)-Ib and aadA1, the trimethoprim-resistance genes dfrA1 and dfrA16, and the β-lactamase bla OXA-2 were found. The obtained MICs in the positive integron isolates to several antibiotics are exposed in Table 3. No clear contribution of gene cassettes could be established in the studied isolates. Only in the strain Ax44, harboring the gene cassette dfrA16, a contribution to the MIC to TMS (256 μg/ml) could be suggested, as it corresponded to the highest value among isolates under scrutiny (Table S1 in Supplementary material).
Schematic representation of arrays of class 1 integrons found among the A. xylosoxidans (n = 24) isolates. Thin black vertical closed bar The attI1 site, thin gray vertical closed bar the attC sites of the gene cassettes. Arrows The primers used to identify the class 1 integron vr. Figure is not in scale
Furthermore, three nosocomial isolates apart from harboring class 1 integrons also have class 2 integrons (Ax79, Ax126, and Ax169) (Table 1). To identify the gene cassette content found in the variable region of class 2 integrons, PCR cartography was performed using different combinations of primers [5, 14, 16]. Only positive amplifications were obtained for the Ax126 showing the presence of the array intI2-sat2-aadA1. The occurrence of the Tn7 transposition gene was also searched, showing that the tnsE gene was present in all isolates, being the tnsB also present in the Ax126 isolate. The rest of the genes gave negative results. To the best of our knowledge, this is the first description of class 2 integrons in Achromobacter spp. [16]. No association of integrons with IS26 and IS440 was found in this study.
In relation with the bla ampC gene previously described in this species [17], it was found in all isolates, confirming its ubiquitous nature.
The exposed results showed that almost all isolates (17/24) included in this study have the capability of carrying ISs, R plasmids, and integrons, associated to horizontal gene transfer usually found in gram-negative clinical isolates. Moreover, the similar GC% between the trfA replicon of the IncP plasmid and the A. xylosoxidans genome reinforces the argument that A. xylosoxidans could be considered as a reservoir of transferable elements. It is likely that its intrinsic antibiotic multidrug resistant profile that ensures its selection under antibiotic pressure, along with its ability to survive in fluids and in the environment [18], makes A. xylosoxidans a reservoir of transferable elements that could contribute to the dissemination and acquisition of antimicrobial resistance mechanisms within the nosocomial environment.
References
Antunes P, Machado J, Peixe L (2007) Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob Agents Chemother 51:1545–1548
Aubert D, Naas T, Nordmann P (2003) IS1999 increases expression of the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. J Bacteriol 185:5314–5319
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228
CLSI (2008) Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA
Flores C, Qadri MI, Lichtenstein C (1990) DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res 18:901–911
Gomez-Cerezo J, Suarez I, Rios JJ, Pena P, Garcia de Miguel MJ, de Jose M, Monteagudo O, Linares P, Barbado-Cano A, Vazquez JJ (2003) Achromobacter xylosoxidans bacteremia: a 10-year analysis of 54 cases. Eur J Clin Microbiol Infect Dis 22:360–363
Hansen CR, Pressler T, Nielsen KG, Jensen PO, Bjarnsholt T, Hoiby N (2010) Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J Cyst Fibros 9:51–58
Magni A, Trancassini M, Varesi P, Iebba V, Curci A, Pecoraro C, Cimino G, Schippa S, Quattrucci S (2010) Achromobacter xylosoxidans genomic characterization and correlation of randomly amplified polymorphic DNA profiles with relevant clinical features [corrected] of cystic fibrosis patients. J Clin Microbiol 48:1035–1039
Merkier AK (2009) Caracterizacion de B-lactamasas en bacilos gram negativos no fermentadores de la glucosa. Thesis Doctoral, Universidad de Buenos Aires
Naas T, Poirel L, Karim A, Nordmann P (1999) Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol Lett 176:411–419
Novais A, Canton R, Valverde A, Machado E, Galan JC, Peixe L, Carattoli A, Baquero F, Coque TM (2006) Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob Agents Chemother 50:2741–2750
Orman BE, Pineiro SA, Arduino S, Galas M, Melano R, Caffer MI, Sordelli DO, Centron D (2002) Evolution of multiresistance in nontyphoid salmonella serovars from 1984 to 1998 in Argentina. Antimicrob Agents Chemother 46:3963–3970
Quiroga MP, Andres P, Petroni A, Soler Bistue AJ, Guerriero L, Vargas LJ, Zorreguieta A, Tokumoto M, Quiroga C, Tolmasky ME, Galas M, Centron D (2007) Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob Agents Chemother 51:4466–4470
Ramirez MS, Quiroga C, Centron D (2005) Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 49:5179–5181
Ramirez MS, Parenteau TR, Centron D, Tolmasky ME (2008) Functional characterization of Tn1331 gene cassettes. J Antimicrob Chemother 62:669–673
Ramirez MS, Pineiro S, Centron D (2010) Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother 54:699–706
Shin KS, Han K, Lee J, Hong SB, Son BR, Youn SJ, Kim J, Shin HS (2005) Imipenem-resistant Achromobacter xylosoxidans carrying blaVIM-2-containing class 1 integron. Diagn Microbiol Infect Dis 53:215–220
Vay CA, Almuzara MN, Rodriguez CH, Pugliese ML, Lorenzo Barba F, Mattera JC, Famiglietti AM (2005) ‘In vitro’ activity of different antimicrobial agents on gram-negative nonfermentative bacilli, excluding Pseudomonas aeruginosa and Acinetobacter spp. Rev Argent Microbiol 37:34–45
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Acknowledgments
M.S.R and D.C. are members of the Carrera del Investigador Científico, CONICET, Argentina. This study was supported by Grant UBACyT 20020100300013 from UBA and PIP 11420100100152 from CONICET to M.S.R., UBACyT M008 and B084 Buenos Aires, Argentina to D.C. and C.V., respectively. C.A. was supported by LA Basin Minority Health and Health Disparities International Research Training Program (MHIRT) 5T37MD001368-14 (National Institute on Minority Health and Health Disparities).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Traglia, G.M., Almuzara, M., Merkier, A.K. et al. Achromobacter xylosoxidans: An Emerging Pathogen Carrying Different Elements Involved in Horizontal Genetic Transfer. Curr Microbiol 65, 673–678 (2012). https://doi.org/10.1007/s00284-012-0213-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-012-0213-5