Abstract
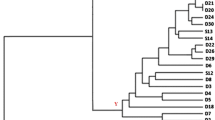
Three molecular typing techniques were applied to assess the molecular relationships of Saccharomyces cerevisiae strains isolated from winery equipment, grapes, and spontaneous fermentation in a cellar located in “Zona Alta del Río Mendoza” (Argentina). In addition, commercial Saccharomyces strains widely used in this region were also included. Interdelta PCR typing, mtDNA restriction analysis, and microsatellite (SSR) genotyping were applied. Dendrograms were constructed based on similarity among different patterns of bands. The combination of the three techniques discriminated 34 strains among the 35 isolates. The results of this study show the complex relationships found at molecular level among the isolates that share the same ecological environment, i.e., the winemaking process. With a few exceptions, the yeast isolates were generally clustered in different ways, depending on the typing technique employed. Three clusters were conserved independently of the molecular method applied. These groups of yeasts always clustered together and had high degree of similarity. Furthermore, the dendrograms mostly showed clusters combining strains from winery and fermentation simultaneously. Most of the commercial strains included in this study were clustered separately from the other isolates analyzed, and just a few of them grouped with the strains mainly isolated from spontaneous fermentation. Only one commercial strain was clustered repetitively with a noncommercial strain isolated from spontaneous fermentation in the three dendrograms. On the other hand, this study has demonstrated the importance of selecting an appropriate molecular method according to the main objectives of the research.




Similar content being viewed by others
References
Ribereau-Gayon P, Dubourdieu D, Donèche B, Lonvaud A (eds) (2006) Cytology, taxonomy and ecology of grape and wine yeast. In: Handbook of enology—the microbiology of wine and vinifications, 2nd edn, vol 1. John Wiley & Sons, Chichester, pp 1–49
Agnolucci M, Scarano S, Santoro S et al (2007) Genetic and phenotypic diversity of autochthonous Saccharomyces spp. strains associated to natural fermentation of ‘Malvasia delle Lipari’. Lett Appl Microbiol 45:657–662
Blanco P, Ramilo A, Cerdeira M, Orriols I (2006) Genetic diversity of wine Saccharomyces cerevisiae strains in an experimental winery from Galicia (NW Spain). Antoine van Leeuwenhoek 89:351–357
Lopes C, Lavalle T, Querol A, Caballero A (2005) Combined use of killer biotype and mtDNA-RFLP patterns in a Patagonian wine Saccharomyces cerevisiae diversity study. Antonie van Leeuwenhoek 3:1–10
Mercado L, Dalcero A, Masuelli R, Combina M (2007) Diversity of Saccharomyces strains on grapes and winery surfaces: Analysis of their contribution to fermentative flora of Malbec wine from Mendoza (Argentina) during two consecutive years. Food Microbiol 24:403–412
Santamaría P, Garijo P, López R et al (2005) Analysis of yeast population during spontaneous alcoholic fermentation: effect of the age of the cellar and the practice of inoculation. Int J Food Microbiol 103:49–56
Torija M, Rozes N, Poblet M et al (2001) Yeast population dynamics in spontaneous fermentations: comparison between two different wine-producing areas over a period of three years. Antonie van Leeuwenhoek 79:345–352
Blondin B, Vezinhet F (1988) Identification de souches de levures oenologiques par leurs caryotypes obtenus en électrophorèse en champ pulsé. Rev Fr Oenol 115:7–11
Querol A, Barrio F, Ramon D (1992) A comparative study of different methods of yeast strain characterization. Syst Appl Microbiol 15:439–446
Legras J, Karst F (2003) Optimisation of interdelta for Saccharomyces cerevisiae strain characterization. FEMS Microbiol Lett 221:249–255
Ness C, Lavalle F, Dubourdieu D et al (1993) Identification of yeast strains using the polymerase chain reaction. J Sci Food Agric 62:89–94
Gallego F, Perez G, Martinez I, Hidalgo P (1998) Microsatellites obtained from database sequences are useful to characterize Saccharomyces cerevisiae. Am J Enol Vitic 49:350–351
Jubany S, Tomasco I, Ponce de León I et al (2008) Toward a global database for the molecular typing of Saccharomyces cerevisiae strains. FEMS Yeast Res 8:472–484
Legras J, Ruh O, Merdinoglu D, Karst F (2005) Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. Int J Food Microbiol 102:73–83
Pérez M, Gallego F, Hidalgo P (2001) Evaluation of molecular techniques for the genetic characterization of Saccharomyces cerevisiae strains. FEMS Microbiol Lett 205:375–378
Martínez C, Cosgaya P, Vásquez C et al (2007) High degree of polymorphism and geographic origin of wine yeast strains. J Appl Microbiol 103:2185–2195
Schuller D, Alves H, Dequin S, Casal M (2005) Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiol Ecol 51:167–177
Schuller D, Valero E, Dequin S, Casal M (2004) Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiol Lett 231:19–26
Combina M, Elía A, Mercado L et al (2005) Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int J Food Microbiol 99:237–243
Hoffman C, Winston F (1987) A ten-minute DNA preparation from yeast efficiently release autonomous plasmids for transformation of E. coli. Gene 57:267–272
Liti G, Carter D, Moses A et al (2009) Population genomics of domestic and wild yeast. Nature 458:337–341
Vigentini I, Fracassetti D, Picozzi C, Foschino R (2009) Polymorphism of Saccharomyces cerevisiae involved in wine production. Curr Microbiol 58:211–218
Carreto L, Eiriz M, Gomes A et al (2008) Comparative genomics of wild type yeast strains unveil important genome diversity. BMC Genomics 9:524
Schacherer J, Shapiro J, Ruderfer D, Kruglyak L (2009) Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458:342–346
Pramateftaki PV, Lanaridis P, Typas MA (2000) Molecular identification of wine yeast at species or strain level: a case study with strains from two vine-growing areas of Greece. J Appl Microbiol 89:236–248
Schuller D, Casal M (2007) The genetic structure of fermentative vineyard-associated Saccharomyces cerevisiae populations revealed by microsatellite analysis. Antonie van Leeuwenhoek 91:137–150
Fernández-Espinar M, López V, Ramón D et al (2001) Study of the authenticity of commercial wine yeast strains by molecular techniques. Int J Food Microbiol 70:1–10
Guillamón JM, Barrio E, Querol A (1996) Characterization of wine yeast strains of the Saccharomyces genus on the basis of molecular markers: relationships between genetic distance and geographic or ecological origin. Syst Appl Microbiol 19:122–132
Puig S, Querol A, Barrio E, Pérez-Ortín JE (2000) Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol 66:2057–2061
Bond U (2009) Chapter 6 the genomes of lager yeasts. Adv Appl Microbiol 69:159–182
Pretorius IS (2000) Tailoring wine yeast for the new millenium: novel approaches to the ancient art of winemaking. Yeast 16:1–55
Rachidi N, Barre P, Blondin B (1999) Multiple Ty-mediated chromosomal translocation lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol Gen Genet 261:841–850
Castrejón F, Codón A, Cubero B, Benítez T (2002) Acetaldehyde and ethanol are responsible for mitochondrial DNA (mtDNA) restriction fragment length polymorphism (RFLP). Syst Appl Microbiol 25:462–467
Acknowledgments
This study was supported by Viticulture Regional Project MZASJ O7 from the Instituto Nacional de Tecnología Agropecuaria (INTA) (and Project PDT 32/06 from Dinacyt, Uruguay). L.M. is a fellow of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Programa de Doctorado en Ciencias Biológicas (PROBIOL), Universidad Nacional de Cuyo.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mercado, L., Jubany, S., Gaggero, C. et al. Molecular Relationships Between Saccharomyces cerevisiae Strains Involved in Winemaking from Mendoza, Argentina. Curr Microbiol 61, 506–514 (2010). https://doi.org/10.1007/s00284-010-9645-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9645-y