Abstract
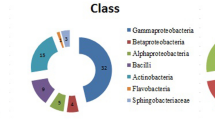
In this study, a total of 130 rhizobacteria was isolated from a saline infested zone of wheat rhizosphere, and screened for plant growth promoting (PGP) traits at higher salt (NaCl) concentrations (2, 4, 6, and 8%). The results revealed that 24 rhizobacterial isolates were tolerant at 8% NaCl. Although all the 24 salt tolerable isolates produced indole-3-acetic acid (IAA), while 10 isolates solubilized phosphorus, eight produced siderophore, and six produced gibberellin. However, only three isolates showed the production of 1-aminocyclopropane-1-carboxylate (ACC) deaminase. Diversity was analyzed through 16S rDNA-RFLP, and of these isolates with three tetra cutter restriction enzymes (HaeIII, AluI, and MspI), the representative cluster groups were identified by 16S rDNA sequencing. Bacillus and Bacillus-derived genera were dominant which showed PGP attributes at 8% NaCl concentration. Out of 24 isolates, nitrogen fixing ability (nif H gene) was detected in the two isolates, SU18 (Arthrobacter sp.) and SU48.





Similar content being viewed by others
References
Ali Z, Salam A, Azhar FM, Khan IA (2007) Genotypic variation in salinity tolerance among spring and winter wheat (Triticum aestivum L.) accessions. S Afr J Bot 73:70–75
Beneduzi A, Peres D, da Costa PB, Zanettini MHB, Passaglia LMP (2008) Genetic and phenotypic diversity of plant-growth-promoting bacilli isolated from wheat fields in southern Brazil. Res Microbiol 159:244–250
Benlloch-Gonzalez M, Fournier J, Ramos J, Benlloch M (2005) Strategies underlying salt tolerance in halophytes are present in Cynara carbunculus. Plant Sci 168:653–659
Borneman J, Skroch PW, O’Sullivan KM (1996) Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol 62:1935–1943
Bradford MM (1976) A rapid and sensitive method for the quantification of micrograms quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Cheng Z, Park E, Glick BR (2007) 1-Aminocyclopropane-1-carboxylate (ACC) deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can J Microbiol 53:912–918
Douka CE, Apostalakis CG, Skarloy VD (1978) Studies of Rhizobium meliloti isolated from salt-affected soil. Ann Appl Biol 88:457–460
Dworkin M, Foster J (1958) Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol 75:592–601
Edwards U, Rogall TH, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Friedberg E, Walker G, Siede W (1995) DNA repair and mutagenesis. American Society for Microbiology, Washington, DC
Gardener BBM (2004) Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Symposium: the nature and application of biocontrol microbes: Bacillus spp. Phytopathology 94: 1252–1258
Glick BR (1995) The enhancement of plant growth by free-living bacteria. Can J Microbiol 4:109–117
Hua SST, Tsai V, Lichens GM, Noma AT (1982) Accumulation of aminoacids in Rhizobium spp. WR 10001 in response to sodium chloride salinity. Appl Environ Microbiol 44:135–140
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, vol 3. Academic Press, New York
Kiewitz C, Tümmler B (2000) Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol 182:3125–3135
Laguerre G, Allard MR, Revoy F, Amarger N (1994) Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR amplifies 16S rRNA genes. Appl Environ Microbiol 60:56–63
Loper JE, Schroth MN (1986) Influence of bacterial sources of indole-3-acetic acid biosynthetic on root elongation of sugar beet. Phytopathology 76:386–389
Maas EV (1986) Salt tolerance of plants. Appl Agric Res 1:12–26
Mayak S, Tirosh T, Glick BR (1999) Effect of wild-type and mutant plant growth promoting rhizobacteria on the rooting of mung bean cuttings. J Plant Growth Regul 18:49–53
Mehta S, Nautiyal CS (2001) An efficient method for qualitative screening of phosphate solubilizing bacteria. Curr Microbiol 43:51–56
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 28:239–250
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Oliver A, Canton R, Campo P et al (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254
Pandey P, Sahgel M, Maheshwari DK, Johri BN (2004) Genetic diversity of rhizobia isolated from medicinal legumes growing in the sub-Himalayan region of Uttaranchal. Curr Sci 86:202–207
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15
Principe A, Alvarez FA, Castro MG, Zachi L, Fischer SE, Mori GB, Jofre E (2007) Biocontrol and PGPR features in native strain isolated from saline soils of Argentina. Curr Microbiol 55:314–322
Rouatt JW, Katznelson H (1961) A study of bacteria on the root surface and in the rhizosphere soil of crop plants. J Appl Bacteriol 24:164–171
Saitou N, Nie M (1987) The Neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Sharan A, Shikha, Darmwal NS, Gaur R (2008) Xanthomonas campestris, a novel stress tolerant, phosphate-solubilizing bacterial strain from saline–alkali soils. World J Microbiol Biotechnol 24:753–759
Sneath PHA, Sokal RR (1973) Numerical taxonomy—the principles and practice of numerical classification. W. H. Freeman, San Francisco
Tripathi AK, Mishra BM, Tripathi P (1998) Salinity stress responses in the plant growth promoting rhizobacteria, Azospirillum spp. J Biosci 23:463–471
Vikram A, Hamzehzarghani H, Alagawadi AR, Krishnaraj PU, Chandrashekar BS (2007) Production of plant growth promoting substances by phosphate solubilizing bacteria isolated from vertisols. J Plant Sci 2:326–333
Yildirim E, Turan M, Donmez MF (2008) Mitigation of salt stress in radish (Raphanus sativus l.) by plant growth promoting rhizobacteria. Roumanian Biotechnol Lett 13:3933–3943
Acknowledgments
This research work was supported by the Department of Environmental Science, Babasaheb Bhimrao Ambedkar University, Lucknow, India and ICAR-Network Project, Indian Council of Agricultural Research, Government of India, New Delhi. Authors are thankful to the funding agencies.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Upadhyay, S.K., Singh, D.P. & Saikia, R. Genetic Diversity of Plant Growth Promoting Rhizobacteria Isolated from Rhizospheric Soil of Wheat Under Saline Condition. Curr Microbiol 59, 489–496 (2009). https://doi.org/10.1007/s00284-009-9464-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9464-1