Abstract
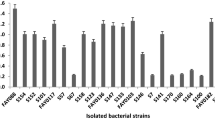
A cellulase-producing bacterium strain was isolated from soil that produced novel thermoalkalotolerant cellulases after growth on CMC-Na agar screening plate at 37°C. It was identified as Escherichia coli using the method of 16S rRNA and intergenic spacer gene analysis combined with morphological, physiological, and biochemical tests. Three major components of the cellulases [carboxymethyl cellulase (CMCase), filter paper cellulase, and β-glucosidase] were produced with maximal activities (0.23, 0.08, and 0.15 U/ml) and maximum specific activities 4.13, 0.56, and 0.50 U/mg protein after 72, 96, and 120 h growth, respectively. Maximum CMCase activity was measured at 50°C and pH 6.0, respectively, and it also retained more than 60% of its maximal activity for at least 20 min at 50–70°C and 10 min at 80°C, respectively, and retained approximately 50% of its maximal activity after incubating at 90°C for 10 min. The enzyme could be applied in bioconversion of lignocellulosic agricultural wastes.




Similar content being viewed by others
References
Thongekkaew J, Ikeda H, Mesaki K, Lefuji H (2008) An acidic and thermostable carboxymethyl cellulase from the yeast Cryptococcus sp. S-2: purification characterization and improvement of its recombinant enzyme production by high cell-density fermentation of Pichia pastoris. Protein Expr Purif 60(2):140–146
Zhang Y, Himmel M, Mielenz J (2006) Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv 24:452–481
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59(2):257–268
Chahal DS (1985) Solid-state fermentation with Trichoderma reesei for cellulase production. Appl Environ Microbiol 49:205–210
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Jang H, Chen K (2003) Production and characterization of thermostable cellulases from Streptomyces transformant T3-1. World J Microbiol Biotechnol 19:263–268
Naimi A, Beck G, Branlant C (1997) Primary and secondary structures of rRNA spacer regions in enterococci. Microbiology 143:823–834
Hinrikson HP, Dutly F, Altwegg M (1999) Homogeneity of 16S–23S ribosomal intergenic spacer regions of Tropheryma whippelii in Swiss patients with Whipple’s disease. J Clin Microbiol 37:152–156
De Wit MYL, Klatser PR (1994) Mycobacterium leprae isolates from different sources have identical sequences of the spacer region between the 16S and 23S ribosomal RNA genes. Microbiology 140:1983–1987
Tanner MA, Goebel BM, Dojka MA, Pace NR (1998) Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol 64:3110–3113
Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Hollien J, Marqusee S (1999) Structural distribution of stability in a thermophilic enzyme. Proc Natl Acad Sci USA 96(24):13674–13678
Te’o VSJ, Saul DJ, Bergquist PL (1995) CelA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl Microbiol Biotechnol 43(2):291–296
Yoshihiro H, Kenzo K, Tadashi Y, Hajimi M, Tohru K, Susumu I (1997) Thermostable alkaline cellulase from an alkaliphilic isolate, Bacillus sp. KSM-S237. Extremophiles 1(3):151–156
Robertson LD, Koehn RD (1978) Characteristics of the cellulase produced by the Ascomycete Poronia punctata. Mycologia 70(5):1113–1121
Zeikus JG, Vieille C, Savchenko A (1998) Thermozymes: biotechnology and structure-function relationships. Extremophiles 2(3):179–183
Acknowledgments
This study was supported by a key project of Zhejiang Government No. 2008C24011 and by the Hi-Tech Research and Development Program of China (Nos. 2008AA10Z132 and 2006AA10A119).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, Xh., Bhaskar, R., Yang, Hj. et al. Screening and Identification of New Isolate: Thermostable Escherichia coli with Novel Thermoalkalotolerant Cellulases. Curr Microbiol 59, 393–399 (2009). https://doi.org/10.1007/s00284-009-9450-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9450-7