Abstract
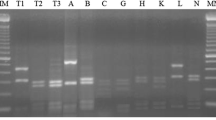
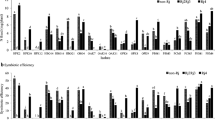
Sixty root nodule isolates of soybean rhizobia indigenous to eight field sites in India were characterized using PCR-RFLP for repeated sequence RSα a 1195-bp DNA fragment, indole acetic acid production, and nitrogenase activity. Site-dependent variations were observed in terms of IAA production and nitrogenase activities. RSα was conserved in slow-growing soybean rhizobia across locations and sites and was absent in other Rhizobiaceae members and other bacterial genera. The results suggest that RSα can be a useful molecular marker for slow-growing soybean rhizobia. The study also showed the low presence of soybean nodulating fast growers in Indian soils.


Similar content being viewed by others
Literature Cited
Gordon AS, Weber RP (1951) Colorimetric estimation of indole acetic acid. Plant Physiol 26:192–195
Hardy RWF, Burns RC, Holstein RD (1968) Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol Biochem 5:47–81
Hartmann A, Gomez M, Giraud JJ, Revellin C (1996) Repeated sequence RS α is diagnostic for Bradyrhizobium japonicum and Bradyrhizobium elkanii. Biol Fertil Soils 23:15–19
Jordan DC (1982) Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. Nov., genus of slow growing and root nodules bacteria from leguminous plants. Int J Syst Bacteriol 32:136–139
Kaluza K, Hahn M, Hennecke H (1985) Repeated sequences similar to insertion elements clustered around the nif region of the Rhizobium japonicum genome. J Bacteriol 162:535–542
Keyser HH, Bohlool BB, Hu TS, Weber DF (1982) Fast growing rhizobia isolated from root nodules of soybean. Science 215:1631–1632
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Intnl J Syst Bacteriol 38:358–361
Kuykendall LD, Saxena B, Devine TE, Udell SE (1992) Short interspersed repetitive DNA sequence in prokaryotic genomes. J Bacteriol 174:4525–4529.
Masterson RV, Prakash RK, Amerly AG (1985) Conservation of symbiotic nitrogen fixation gene sequences in R. japonicum and B. japonicum. J Bacteriol 163:21–26
Minamisawa K (1990) Division of rhizobitoxin producing and hydrogen uptake-positive strains of Bradyrhizobium japonicum by nif DKE sequence divergence. Plant Cell Physiol 31:81–89
Minamisawa K, Fukai K (1991) Production of indole acetic acid by Bradyrhizobium japonicum. A correlation with genotype grouping and rhizobitoxine production. Plant Cell Physiol 32:1–9
Minamisawa K, Seki T, Ondera S, Kubota M, Asami T (1992) Genetic relatedness of Bradyrhizobium japonicum field isolates as revealed by repeated sequences and various other characteristics. Appl Environ Microbiol 58:2832–2839
Minamisawa K, Nakatsuka Y, Isawa T (1999) Diversity and field site variation of indigenous populations of soybean bradyrhizobia in Japan by fingerprints with repeated sequences RSα and RSβ . FEMS Microbiol Ecol 29:171–178
SatyaPrakash C, Annapurna K (2006) Diversity of soybean bradyrhizobial population adapted to an Indian soil. J Plant Biochem Biotechnol 15:27–32
Scholla MH, Elkan GH (1984) Rhizobium fredii sp. nov. a fast growing bacterium that effectively nodulates soybeans. Int J Syst Bacteriol 34:484–486
Vincent JM (1970) A manual for the practical study of Root Nodule Bacteria. International Biological Program Handbook No. 15. Oxford, Blackwell Science Ltd. pp 73–97
Wheatcroft R, Laberge S (1991) Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol 173:2530–2538
Wilson KJ, Sessitsch A, Corbo JC, Galler KE, Akkermans ADL, Jefferson RA, (1995) β-glucuronidase (GUS) transposons for ecological studies of rhizobia and other Gram negative bacteria. Microbiol 141:1691–1705
Xu LM, Ge C, Cui Z, Li J, Fan H (1995) Bradyrhizobium liaoningensis sp. nov. isolated from the root nodules of soybeans. Int J Syst Bacteriol 45:706–711
Acknowledgments
Thanks go to the National Phytotron Facility, IARI, for conducting the nodulation tests. B.N is indebted to IARI, New Delhi, for a junior research fellowship
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Annapurna, K., Balakrishnan, N. & Vital, L. Verification and Rapid Identification of Soybean Rhizobia in Indian Soils. Curr Microbiol 54, 287–291 (2007). https://doi.org/10.1007/s00284-006-0423-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0423-9