Abstract
Autophagy, a catabolic process involved in the sequestration and lysosomal degradation of cytoplasmic contents, is crucial for cellular homeostasis. The current literature supports that autophagy plays diverse roles in the development, maintenance, and progression of tumors. While genetic evidence indicates autophagy functions as a tumor suppressor mechanism, it is also apparent that autophagy can promote the survival of established tumors under stress conditions and in response to chemotherapy. In this review, we discuss the mechanisms and the evidence underlying these multifaceted roles of autophagy in tumorigenesis, the prospects for targeting autophagy in cancer therapy, and overview the potential markers that may be utilized to reliably detect autophagy in clinical settings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Macroautophagy (autophagy) is a tightly regulated self-digestive process in which cytoplasmic contents and organelles are sequestered within double-membrane vesicles, called autophagosomes, and delivered to the lysosome for degradation and recycling. Autophagy is regulated by a limited number of highly conserved genes called ATGs (for AuTophaGy gene). Although originally identified in yeast [1], there have been numerous recent breakthroughs in mammals demonstrating how autophagy critically regulates key physiological and pathological processes. Autophagy serves vital housekeeping functions in cells, including the turnover of damaged organelles and long-lived proteins [2]. Furthermore, the bulk degradation of cellular material through autophagy allows cells to recycle both nutrients and energy during starvation and stress [3]. Thus, autophagy is proposed to function as a “battery” that buys cells valuable time, allowing them to survive if the stressor is removed in a timely manner. The indispensable contribution of cellular autophagy to physiological homeostasis has been demonstrated by studies in which mice with genetic deletion of critical ATGs die within a day after birth [4, 5]. On the other hand, an excessive level of autophagy has been proposed to promote cell death due to the over-consumption of critical cellular constituents [6]; this process has been termed ATG-dependent or type II-programmed cell death.
Overall, the malfunctioning of this cellular self-eating has been linked to a wide range of pathological conditions including neurodegenerative diseases, pathogen infection, and cancer [7]. Autophagy is activated in response to multiple stresses during cancer progression, such as nutrient starvation, the unfolded protein response (UPR) (endoplasmic reticulum (ER) stress), and hypoxia; in addition, it is observed upon treatment of cancers with a wide spectrum of cytotoxic and targeted chemotherapeutic agents [8, 9]. In this review, we will summarize the current understanding of the role of autophagy in tumorigenesis, the current status and opportunities of autophagy manipulation in cancer therapy and also discuss the need for identification of suitable markers of autophagy in clinical settings.
The molecular regulation of autophagy
Autophagosome formation
Autophagy is a multi-step process characterized by the induction, nucleation, extension, and completion of an isolation membrane (also termed the phagophore) to form an organelle called the autophagosome; cytosolic proteins and organelles sequestered within the autophagosome are subsequently delivered to the lysosome for degradation (Fig. 1). Studies conducted in yeast have revealed over 30 ATG genes involved in this process, many of whose mammalian orthologues have also been identified.
Schematic depiction of the autophagy pathway. Autophagy is a multi-step process characterized by the induction, nucleation, extension, and completion of an isolation membrane (phagophore). The initiation of autophagy is regulated by the ULK complex. Nucleation of the phagophore is regulated by the Class III PI3K complex, which consists of Vps34, Beclin1, p150, and ATG14. Membrane elongation requires two ubiquitin-like systems that mediate the formation of ATG12-ATG5 and PE-conjugated ATG8. The completed autophagosome, along with sequestered cargo, ultimately fuses with the lysosome. The resulting autolysosome is a single membrane-bound acidic vesicle where the contents are digested by lysosomal hydrolases and recycled. Further details are provided in the text
ULK complex
In mammals, the induction of autophagy requires the unc-51-like kinase (ULK; ATG1 in yeast), which exists in a large complex with mATG13 and FIP200 that is regulated by mammalian target of rapamycin (mTOR) [10]. Although it bears no structural homology to the yeast counterpart, FIP200 has been proposed to be a functional orthologue of yeast ATG17 during autophagosome induction [11]. Interestingly, truncation mutants of this protein have been described in breast cancer patients [12].
There are at least three different ULK proteins involved in different aspects of autophagy. However, ULK1 and ULK2 bear highest similarity to yeast ATG1. Under nutrient rich conditions, the ULK complex interacts with mTORC1 and remains inactivated by mTORC1-mediated phosphorylation of ULK1 and ULK2. However, upon nutrient deprivation, mTORC1 dissociates from the complex causing dephosphorylation and activation of ULK1 and ULK2, which can then phosphorylate and activate mATG13 and FIP200, leading to subsequent localization of the activated ULK complex to the phagophore [13]. However, the substrates of the ULK complex and how it regulates the autophagic machinery are not entirely known. Remarkably, the genetic deletion of ULK1 in mice has been demonstrated to have minimal effects on the initial formation or completion of the autophagosome; this lack of phenotype may arise from compensation by other ULK isoforms [14]. Instead, cells lacking ULK1 demonstrate an increase in mitochondrial mass, resulting from the defective clearance of mitochondria via autophagy [14].
Class III phosphatidylinositol 3-kinase complex
A Class III phosphatidylinositol 3-kinase (PI3K) complex is necessary for the formation of phosphatidylinositol 3-phosphate (PI3P), which is essential for the early stages of nucleation of the phagophore. The PI3K core complex consists of the PI3K protein Vps34, beclin1, and p150 [15]. Though the exact mechanism is not yet clear, it is evident that the core complex localizes to the phagophore and facilitates recruitment of subsequent ATGs. Recent studies have identified various binding partners of beclin1, including ultraviolet (UV) radiation resistance-associated gene (UVRAG) [16, 17], ATG14L/Barkor [18, 19], and Ambra1 [20] which all positively regulate beclin1 activity and regulate different steps of autophagosome formation and maturation. Moreover, another molecule named Rubicon (RUN domain and cysteine-rich domain containing, Beclin1-interacting protein) has been uncovered as a negative regulator of beclin1 [18], which binds to the UVRAG–beclin complex and regulates late stages of autophagy, more specifically, the late endosomal and lysosomal maturation process.
Ubiquitin-like conjugation pathways
The next stage of phagophore membrane elongation requires two ubiquitin-like systems [21, 22]. The first involves the conjugation of ATG5 to ubiquitin-like ATG12 via E1- and E2-like activities of ATG7 and ATG10, respectively. The ATG5–ATG12 complex binds ATG16 and forms a large multimeric complex called the ATG16L complex, which localizes on the outer surface of the extending autophagosomal membrane. The second conjugation system involves cleavage of the ubiquitin-like molecule, ATG8, by the protease ATG4 to expose a C-terminal glycine residue required for subsequent activation and conjugation reactions. Ultimately, ATG8 is conjugated to the lipid phosphotidylethanolamine (PE) via ATG7 and E2-like ATG3, and is subsequently recruited to both the outer and inner surfaces of the autophagosomal membrane. Several mammalian orthologues to ATG8 have been identified, of which the best characterized is microtubule-associated protein light chain 3 (MAPLC3 or LC3) [23]. Importantly, the ATG16L complex with ATG5–ATG12 has been demonstrated to serve as an E3 that promotes the lipidation of ATG8/LC3 [24, 25]. Moreover, recent evidence suggests that ATG8/LC3 conjugation system can also regulate the ATG16L complex because in ATG3-deficient mice, where no detectable LC3 lipidation is present, there is marked impairment of ATG5-ATG12 conjugation [26]. To date, these conjugation reactions have been proposed to be uniquely devoted to the process of autophagosome formation, presumably by promoting membrane fusion and biogenesis during the elongation of the phagophore; as a result, they have been viewed as specific drug targets to potentially modulate autophagy in cancer [27].
Autophagosome maturation
The completion of the autophagosome membrane is followed by its subsequent fusion with the lysosome via the action of the lysosomal proteins LAMP1 and LAMP2, as well as Rab7 and UVRAG [28]. The resulting autolysosome is a single membrane-bound acidic vesicle where the contents are digested and recycled by lysosomal hydrolases (e.g., cathepsins). These single-membrane autolysosomes, filled with degraded cytoplasmic material, are most commonly observed by electron microscopy following the induction of autophagy in response to diverse stimuli.
Regulation of autophagy by mammalian target of rapamycin
Significant progress has been made over the past years in determining the molecular regulators of autophagy. For detailed understanding of autophagic regulators, additional reading is recommended [29]. Here, we principally focus on the role of the mTOR pathway in autophagy regulation.
The serine/threonine kinase mTOR plays a pivotal role in maintaining the balance between cell growth and autophagy in response to growth factor signaling, nutrient status, and stress. mTOR exists in two complexes: mTORC1 (consisting of mTOR, GβL, raptor, Deptor, PRAS40), which is sensitive to rapamycin, and mTORC2 (consisting of mTOR, GβL, rictor, Sin1, PRR5, Deptor) [30]. Activated mTORC1 promotes mRNA translation by activating S6K and inhibiting 4EBP1; in addition, it also inhibits autophagy by phosphorylating the ATG13-ULK-FIP200 complex [31]. Regulation of mTORC1 activity in response to diverse stimuli is primarily regulated by the tuberous sclerosis complex (TSC) proteins [32]. TSC2, in a complex with TSC1, acts as a GTPase activating protein for the small GTPase Ras homolog enriched in brain (Rheb) that activates mTORC1 in its GTP-bound form [33].
The role of mTOR in mediating growth factor signaling is primarily in response to the class I PI3K-protein kinase B (Akt) pathway [34]. Activated growth factor receptor stimulates PI3K to convert phosphotidylinositol 4,5-bisphosphate to phosphotidylinositol 3,4,5-trisphosphate, leading to the recruitment of phosphatidylinositide-dependent protein kinase 1 (PDK1) and Akt to the plasma membrane followed by phosphorylation and activation of Akt by PDK1. Activated Akt then phosphorylates TSC2 and prevents TSC complex formation and thus leads to mTOR activation. Though the role of mTORC2 in autophagy is not clearly delineated, it is important to recognize that mTORC2 activates Akt; because Akt positively regulates mTORC1 activity, one can speculate that mTORC2 also negatively regulates autophagy [35]. During tumorigenesis, constitutive activation of the PI3K pathway often leads to increased mTORC1 activation, presumably resulting in autophagy inhibition. In fact, inhibition of Akt kinase activity has been recently shown to induce autophagy levels [36]. On the other hand, overexpression of tumor suppressor phosphatase and tensin homolog (PTEN), the negative regulator of PI3K/AKT pathway, induces autophagy [37].
mTOR is also a critical sensor of cellular energy and nutrient status. Reduced ATP production during nutrient deprivation results in an elevated AMP/ATP ratio and activates the energy-sensing serine/threonine kinase 11 (LKB1)-adenosine monophosphate–activated protein kinase (AMPK) signaling axis. AMPK mediates the phosphorylation of TSC, leading to inactivation of mTORC1 and induction of autophagy [38]. AMPK can also directly phosphorylate Raptor and thus regulate mTORC1 in a TSC-independent manner [39]. Moreover, the cellular amino acid pool, especially branched chain amino acids, critically regulates mTOR activity. A recent study revealed that cellular uptake of L-glutamine by the transporter SLC1A5 followed by its rapid efflux via the bidirectional transporter SLC7A5/SLC3A2, which is responsible for simultaneous L-glutamine efflux and leucine import, results in mTORC1 activation. Thus, loss of SLC1A5 inhibits cell growth and activates autophagy presumably due to inhibition of leucine uptake into cells [40].
Finally, mTOR also serves as an important sensor of cellular stress. Regulated in development and DNA damage response 1 (REDD1) is induced by hypoxia and leads to mTORC1 inhibition by regulating the TSC complex [41]. Moreover, the tumor suppressor protein p53, which is induced in response to diverse genotoxic stress, has also been shown to promote transcription of various negative regulators of the mTOR pathway, such as AMPKβ, TSC2, and PTEN [42, 43]. Furthermore, sestrin, which is induced in response to DNA damage and oxidative stress in a p53-dependent manner, was recently shown to inhibit mTORC1 activity via AMPK activation [44]. Additional studies revealed that sestrin2 is indeed required for autophagy induction in response to various stress inducers, including nutrient starvation and rapamycin [45], thereby further establishing the link between p53, mTORC1, and autophagy. However, it is important to recognize that the role of p53 in autophagy regulation is not straightforward, because it can also promote autophagy in an mTOR-independent manner via the transcriptional upregulation of its downstream target damage-regulated autophagy modulator (DRAM) [46].
Though additional pathways exist, mTOR, which is positioned at the crossroads of various critical signaling pathways, serves as a focal point of autophagy regulation. Deregulation of mTOR activity due to perturbation in upstream signaling events during carcinogenesis can therefore have significant impact on autophagy levels and subsequent tumor outcome. Moreover, mTOR regulation of autophagy may significantly impact the efficacy of the growing number of anti-cancer agents targeting the PI3K/mTOR pathway [47].
Role of autophagy in cancer: context matters
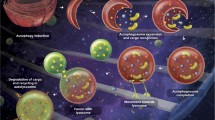
It is increasingly appreciated that the contribution of autophagy to cancer is extremely complex. There is ample evidence supporting both tumor promoting and suppressive functions of autophagy. This apparent paradox can be reconciled by the fact that the distinct role(s) of autophagy during cancer progression is/are tumor type, context and stage specific (Fig. 2). While genetic evidence suggests the tumor suppressive functions of autophagy act during tumor initiation, there is also compelling evidence that autophagy is used as a survival strategy by established tumors to cope with diverse stresses of the microenvironment that are encountered during tumor progression and metastasis. Overall, the importance of autophagy in carcinogenesis, as well as its potential modulation in cancer therapy, is gaining significant attention. Autophagy levels, as well as expression of autophagic molecules, are being used as prognostic markers for predicting tumor outcome [48–50].
Schematic depiction of the tumor suppressive and pro-tumorigenic roles of autophagy during cancer progression and therapy. Tumor suppressive functions of autophagy include: (1) eliminating damaged organelles and mitigating oxidative stress, which prevents genome instability, and ultimately, malignant transformation; (2) promoting oncogene-induced senescence, a barrier to malignant transformation; and (3) preventing necrosis in apoptosis deficient cells in response to metabolic stress, which reduces pro-tumorigenic inflammation. Pro-tumorigenic roles for autophagy include: (1) promoting tumor cell survival in response to diverse micro-enviromental and chemotherapy-induced stresses, and (2) protecting cells from anoikis, which may facilitate drug-resistance and metastasis. Further details are provided in the text
Autophagy in tumor suppression
Since the prototypic functions of autophagy are to recycle essential nutrients and provide energy for survival during starvation and stress, it initially seems counterintuitive that autophagy can act as a potential tumor suppressor mechanism. However, genetic evidence supports that autophagy can prevent tumor formation. The role of autophagy as a tumor suppressor was first broached through genetic studies of beclin1, the mammalian orthologue of yeast ATG6 [51]. Beclin1 was mapped to a tumor susceptibility locus that is monoallelically deleted in a high percentage of human breast, ovarian, and prostate cancers; furthermore, analysis of human tissue samples revealed decreased beclin1 expression in human breast carcinomas compared to normal breast tissue [51]. Moreover, oncogenes like Akt [36], Ras, and ERK [52] inhibit autophagy primarily by activating the mTOR signaling pathway. On the other hand, there is abundant evidence that tumor suppressors like p53 [44, 46], PTEN [37], and ARF [53, 54] activate autophagy.
Genetic evidence for tumor suppressor functions by autophagy-related genes
Beclin1
Beclin1, the mammalian orthologue to yeast ATG6, was identified due to its interaction with Bcl-2 [55, 56]. Bcl-2 binds to beclin1 and inhibits its autophagic activity by blocking its interaction with Vps34. Genetic studies revealed that mice homozygously deleted for beclin1 die during embryogenesis [57]. Nonetheless, two groups demonstrated that heterozygous disruption of beclin1 promoted tumorigenesis in mice, albeit over an extended latency. Mice lacking a single copy of beclin1 (beclin +/−) developed spontaneous tumors, including lymphoma, hepatocellular carcinoma, lung adenocarcinomas, and mammary hyperplasia [57, 58]. Notably, the loss of the second allele of beclin did not occur in these tumors, which strikingly resembled the monoallelic losses originally found in human patients. Thus, these reports provided the first direct genetic evidence that beclin1/atg6 functions as a haploinsufficient tumor suppressor. Moreover, ectopic overexpression of beclin1 in MCF7 mammary carcinoma cells, which exhibits low beclin1 expression, causes reduced cancer cell proliferation in vitro and decreased tumorigenic potential in vivo, further suggesting a role for this autophagy regulator in tumor suppression [51].
UV irradiation resistance-associated gene
UVRAG is a beclin1-interacting protein that positively regulates autophagy [16]. Like beclin1, UVRAG is monoallelically deleted in human colon carcinoma [16]. Moreover, frameshift mutations in the polyadenine tract of the UVRAG gene are present in gastric carcinomas, and the gastric cancer cells with the mutations have reduced autophagy activity [59].
Bif1
Originally identified as a Bax binding protein, Bif1 was recently shown to interact with beclin1 through UVRAG. It promotes autophagosome membrane curvature via its N-terminal BAR (Bin/Amphiphysin/Rvs) domain and thus plays a crucial role in isolation membrane extension. Bif 1 knockout mice have significantly higher rate of spontaneous tumors [60] and reduced Bif1 expression is observed in gastric carcinoma [61].
Other ATG genes
ATG4c knockout mice are viable after birth and do not show higher rate of spontaneous tumors compared to wild-type animals. However, they have a higher susceptibility for formation of fibrosarcomas when exposed to chemical carcinogens [62]. Frameshift mutations in ATG2B, ATG5, and ATG9B were recently reported in gastric and colorectal carcinomas with high microsatellite instability, further underlining the tumor suppressor function of the core autophagic machinery [63].
Mechanisms of tumor suppression by autophagy
Autophagy prevents the accumulation of damaged organelles, oxidative stress, inflammation, and genomic instability
A mounting body of work from Eileen White and colleagues demonstrates an important role for autophagy in protecting cells from genotoxic stress and maintaining genome integrity. This specific tumor suppressor function of autophagy has thus far been best characterized in the context of metabolic stress, a condition typically observed in tumors due to hypoxia and inadequate glucose supply coupled with increased energy demands of the rapidly proliferating cells, which leads to extensive cellular damage. Apoptosis acts as the first line of defense to remove these damaged cells. However, when apoptosis is reduced or inactivated, which commonly occurs during tumorigenesis, the cells rely significantly on autophagy for ATP maintenance and cellular fitness. In Bcl-2-overexpressing immortalized mouse mammary epithelial cells and immortalized baby mouse kidney epithelial cells, loss of one copy of beclin1 significantly sensitizes cells to metabolic stress [64, 65]. Paradoxically, in spite of increased survival, beclin1+/+ cells are less tumorigenic than beclin1+/− cells. This intriguing outcome is due to the fact that in autophagy-defective cells, metabolic stress induced significantly higher DNA double-strand breaks and gene amplification, as well as the accumulation of damaged mitochondria and ER chaperones, compared to their wild-type counterparts. Moreover, p62, a scaffolding protein involved in signal transduction events, as well as directing polyubiquitinated proteins and aggregates to autophagosomal degradation, serves as a critical link between defective autophagy and tumorigenesis [66]. The aberrant accumulation of damaged mitochondria and protein aggregates in autophagy-defective cells leads to elevated reactive oxygen species (ROS) levels, which causes DNA damage as well as p62 accumulation. Again p62 accumulation upon metabolic stress leads to ROS generation, thereby creating a positive feedback loop. Thus, by keeping in check the intracellular ROS levels, autophagy serves as a tumor suppressor function in a cell-autonomous manner. Importantly, one must recognize that these experiments have been conducted in cells harboring multiple genetic abnormalities, including the inactivation of the tumor suppressor p53, which is well-known for its ability to maintain genomic integrity [67]. Hence, these results are most consistent with the hypothesis that defective autophagy functions as a secondary modifier, rather than a fundamental driver, of genomic damage during tumor progression.
Another potential tumor-suppression mechanism of autophagy is via inhibition of necrotic cell death of apoptosis-resistant cells during metabolic stress. Though decreased viability in the absence of autophagy and apoptosis should negatively affect tumorigenicity, Degenhardt et al. showed that the necrotic cell death causes macrophage infiltration and proinflammatory cytokine production and thereby facilitates tumor growth [68]. Remarkably, inflammatory cells infiltrate tumor sites in response to necrosis resulting from hypoxia and metabolic stress, both of which commonly affect solid tumors. Although certain inflammatory cells, such as cytotoxic T cells and NK cells are anti-metastatic, chronic tumor inflammation associated with severe hypoxia and metabolic stress generally favors pro-tumor immunity [69, 70]. Importantly, infiltration of pro-tumor inflammatory mediators, like macrophages, correlates with poor clinical prognosis [71], underscoring the importance of understanding the biological mechanisms by which tumor cells tip the balance in favor of pro-tumor immunity over tumor suppressive immunity. Thus, by limiting tumor cell necrosis, autophagy may serve as a non-autonomous mechanism for tumor suppression by preventing leukocyte infiltration of the primary tumor site.
Autophagy and senescence
Oncogene-induced senescence (OIS), a state of premature permanent cell cycle arrest, is the first major barrier towards cellular transformation in response to the mitotic burst induced by oncogenes. It is characterized by an initial mitotic phase followed by a transition phase and finally the cell cycle arrest. Recent studies have identified that the induction and maintenance of OIS is mediated by an inflammatory network comprised of IL-6 and IL-8 which function in a cell-autonomous manner [72]. Interestingly, Young et al. showed that autophagy is induced during the transition phase of OIS in an inducible Ras cell culture system [73]. The importance of autophagy as an essential OIS effector was established by studies in which significant bypass of senescence and inhibition of secretion of IL6 and IL8 was observed upon ATG5 or ATG7 knockdown. Mechanistically, intracellular recycling associated with autophagy might provide the amino acids for the synthesis of the secretory proteins and thereby facilitate OIS. Thus a basal level of autophagy plays an important role in restricting cell growth and proliferation during oncogenic stress and potentially precludes further genomic insults.
Tumor promoting functions for autophagy
Although reduced autophagy is believed to promote tumor development, a minimal level is believed to be necessary for the survival of cancer cells. Moreover, increased autophagy is observed in transformed cells when exposed to diverse stress. Thus, it is increasingly appreciated that autophagy provides cancer cells with certain selective advantages to cope with stress, both in the primary tumor microenvironment as well as during dissemination and metastasis. In the following section, we overview three scenarios in which autophagy-mediated cell survival promotes tumor progression.
Hypoxia
Tumor hypoxia, resulting from inadequate tumor vasculature, is associated with a more malignant phenotype, higher predisposition for metastasis, and poor prognosis. Hypoxic stress selects for cells that are resistant to apoptosis, as well as poses a major barrier to chemotherapy and radiotherapy. Degenhardt et al. first showed that autophagy is induced specifically in the hypoxic core of tumors where it promotes survival [68]. Further studies have unveiled the molecular connections between hypoxia and the induction of autophagy. Hypoxia-inducible factor 1α (HIF-1α), a key transcription factor encoding a plethora of genes responsible for altered metabolism, angiogenesis, invasion, metastasis, therapy-resistance in hypoxic tumors [74], is a positive regulator of autophagy. Bcl-2/adenovirus E1B 19 kDa-interacting protein (BNIP3), a BH3-only protein, is a downstream target of HIF-1α and was recently shown to induce mitophagy and thereby control ROS production in response to hypoxia [75]. Further mechanistic studies by Bellot et al. showed that induction of BNIP3 and BNIP3L in hypoxic cells disrupts the beclin1-Bcl-2 complex, thereby releasing beclin1 to induce autophagy [76]. Though induction of BNIP3, a proapoptotic protein, was initially implicated in driving autophagic cell death [77], more elaborate studies have clearly revealed that BNIP3-induced autophagy is an adaptive survival response during prolonged hypoxia [76]. In addition, there is emerging evidence that various HIF-1α-independent cellular stress response pathways, like AMPK and UPR, can also mediate hypoxia-induced autophagy [78, 79].
As the role of autophagy as a key mediator of survival of hypoxic cells is emerging, the exact mechanisms underlying this phenotype remain unclear. Because chronic hypoxia leads to major metabolic perturbations in tumor tissues, one can postulate that by recycling basic cellular components, autophagy helps stressed cells cope with the increased metabolic demand. Nonetheless, further studies are needed to validate this hypothesis and unveil the interconnections between hypoxia-driven tumor metabolism and autophagy.
Extracellular matrix detachment
Anoikis, or detachment-induced cell death, serves the homeostatic function of killing cells that have lost contact with the basement membrane. Fung et al. showed that autophagy is induced in non-transformed cells following matrix detachment which protects cells from anoikis [80]. These experiments also delineated that autophagy is specifically induced upon β1 integrin blockade. Moreover, in a three-dimensional (3D) epithelial culture system, autophagy was significantly induced in the detached luminal cells and the knockdown of key autophagy genes resulted in accelerated luminal clearance. These elaborate in vitro studies proves that autophagy is instrumental in anoikis resistance, a process exploited by disseminating tumor cells to survive after detachment from the primary site as well as while migrating to distant metastatic sites. Though an interesting hypothesis, the ability of autophagy to promote tumor dissemination and metastasis by protecting cells from anoikis must still be established in relevant in vivo models.
Tumor dormancy
After initial regression of the primary tumor following therapy, cancer can relapse after a long disease-free period. This occurs due to a small population of tumor cells that escape therapy and exist as micrometastatic cells without any clinical manifestation. Though viable, these cells are not proliferative and are thus resistant to conventional chemotherapy that typically targets rapidly growing cells [81]. Thus, a better understanding of the molecular events leading to tumor dormancy, as well as the development of clinically relevant in vivo models are imperative for identifying suitable treatment options. Recently, autophagy was shown to be crucial for the survival of dormant cells in an ovarian cancer model [82]. Lu et al. showed that autophagy supported the survival of a subset of cells in xenograft ovarian tumors in the face of the cytotoxic effects of the tumor suppressor aplasia Ras homolog member I (ARHI). Upon return of more favorable conditions (i.e. ARHI suppression as observed in clinical ovarian carcinoma), the tumor regained proliferative potential and led to rapid regrowth. Apart from this study, there is no direct evidence for the role of autophagy in tumor dormancy. Nonetheless, there are numerous scenarios in which autophagy may promote dormancy. For instance, tumor dormancy is postulated to be a stress management mechanism adopted by disseminated tumor cells to cope with the unfavorable microenvironment by completely withdrawing from the cell cycle [81]. p27Kip1, the cyclin dependent kinase inhibitor involved in G0/G1 cell cycle arrest, was recently identified as a downstream target of the energy-sensing LKB1–AMPK pathway and was shown to induce autophagy and facilitate cell survival in response to growth factor withdrawal and metabolic stress [83]. Thus, disseminated tumor cells (DTC) may depend on p27-mediated autophagy for survival in an inhospitable microenvironment and to resist chemotherapy. Moreover, studies in breast cancer models suggest that decreased mitogenic signaling resulting from impaired integrin and growth factor signaling may lead to tumor dormancy [84, 85]. As mentioned above, integrin-signaling blockade is a potent inducer of autophagy in matrix-detached cells [80], and thus it can be hypothesized that disrupted integrin signaling-mediated autophagy induction in DTC can support and maintain tumor dormancy.
Manipulating autophagy for cancer therapy
Because autophagy plays a crucial role in tumor growth, one can predict that manipulation of autophagy levels will have significant effects on tumor outcome. A growing number of studies in preclinical settings indicate that both the cytoprotective as well as death-inducing properties of autophagy may be exploited for therapeutic purposes.
Autophagy inhibition as a potential adjuvant therapy in cancer
As discussed above, autophagy is a stress adaptive response that is upregulated by microenvironmental insults, including metabolic stress, growth factor withdrawal, and cellular damage. Similarly, high levels of autophagy are commonly observed in tumor cells following anti-cancer treatment. Specifically, anti-neoplastic therapies, including radiation therapy, chemotherapy (e.g., doxorubicin, temozolomide, camptothecin), histone deacetylase inhibitors, hormonal therapy (e.g., tamoxifen), targeted anti-cancer agents (imatinib, trastuzumab) have all been observed to induce autophagy in various human cancer cell lines (Table 1). In these contexts, autophagy has been proposed to be an Achilles heel in cancer therapy, whose disruption can be a promising strategy to reestablish or increase sensitivity to therapeutic agents. Indeed, ample evidence supports this notion in multiple tumor types and in response to diverse chemotherapeutic agents, highlighting the possibility of targeting autophagy as adjuvant therapy for cancer. In fact, the effect of inhibiting autophagy in combination with current anti-cancer therapies has been tested in multiple tumor models [86].
Pharmacologic inhibition of autophagy
The significant promise of autophagy inhibition in improving clinical efficacy of chemotherapeutic agents calls for the identification and characterization of pharmacological autophagic inhibitors. Chloroquine (CQ), a well-known anti-malarial agent, inhibits lysosomal acidification and blocks the terminal stages of autophagic proteolysis. Remarkably, its cancer preventive potential was discovered decades ago when it was shown to reduce the risk of Burkitt's lymphoma in patients treated for malaria [87]. Currently, the autophagy inhibitory role of CQ and its analogs is being avidly investigated for its potential as a cancer therapeutic. Elegant studies by Amaravadi et al. showed that in a Myc-induced mouse model of lymphoma, CQ and hydroxychloroquine (HCQ) treatment significantly enhanced the cytotoxic effects of p53 expression and alkylating agents [88]. In another study, CQ inhibited spontaneous lymphomagenesis in Eμ-Myc mice, a model for Burkitt's lymphoma, in a p53-dependent manner [89]. While CQ induced apoptosis in Atg7-deficient Myc-overexpressing MEFs in a p53-dependent manner, in apotosis-deficient cells, CQ could induce caspase-independent cell death. Remarkably, simultaneous inhibition of autophagy and apoptosis attenuated the cytotoxic effect of CQ, underscoring the prosurvival role of autophagy in this context. The promising role of CQ as an adjuvant to targeted therapy has also been reported. In chronic myelocytic leukemia (CML) cell lines, inhibition of autophagy by CQ potentiates TKI (imantinib)-mediated cell death [90]. Also, CQ promoted the cytotoxic effects of SAHA in both imatinib-sensitive and -resistant cell lines of CML [91].
These studies and the established clinical use of CQ have prompted clinical trials aimed at investigating the efficacy of CQ in combination with standard cancer therapies for a variety of cancers. However, it must be remembered that the cytotoxic effects of CQ and HCQ might not be solely due to autophagy inhibition [89]. Thus, development of autophagy-targeted agents (e.g. ULK1 inhibitors) might be an attractive strategy for specific autophagy inhibition in cancer therapy. However, such an approach comes with the caveat of inhibiting the homeostatic functions of autophagy in non-transformed cells.
Autophagy-dependent cell death
Autophagy-dependent cell death, also termed type II-programmed cell death, has been described to be an active death mechanism, where extensive degradation of cytoplasmic materials beyond a critical point is believed to drive cell death. However, as the literature supporting the stress adaptive prosurvival role of autophagy rapidly expands, it remains controversial whether increased autophagy observed during a death process serves as an active death-inducing mechanism versus a failed attempt by the cell to survive a specific stress or insult [92]. The potential positive and negative feedback regulation of autophagy and apoptosis further complicates interpretation. Moreover, the lack of in vivo validation of cell culture studies showing autophagy-induced cell death also raises questions with regard to the physiological relevance of such a process. Nonetheless, recent studies do show that autophagy is required for the efficient killing of tumor cells in certain circumstances (Table 1). Here, we discuss some of the notable studies describing autophagy-dependent cell death induced by anti-neoplastic agents in cancer models typically associated with poor treatment outcome. Most importantly, these studies largely support a model in which early and persistent autophagy induction by a particular agent is required for the subsequent induction of classical apoptotic pathways.
First, Salazar et al. showed that Δ9-tetrahydrocannabinol (THC)-mediated ER stress leads to autophagy-dependent cell death in glioma cell lines [93]. In this study, THC treatment induced ER stress and inhibited the AKT/mTOR pathway. Interestingly, autophagy-induced cell death, both in vitro and in vivo, was mediated by apoptosis since cells treated with caspase inhibitors showed the same degree of death resistance as atg5−/− cells and genetic deletion of atg5 or RNAi-mediated depletion of atg1 partially protected cells from apoptosis during THC treatment. More recently Pardo et al. reported that gemcitabine induced autophagy and apoptosis in human pancreatic cancer cell lines. Pharmacological inhibition of autophagy using 3-MA-impaired gemcitabine induced apoptosis, suggesting that autophagy promotes cell death via apoptosis [94].
Finally, upon screening for commercially available immunomodulators for melanoma treatment, Tormo et al. showed that the double-stranded RNA (dsRNA) mimic, polyinosine-polycytidylic acid complexed with polyethyleneimine ([PIC]PEI), is a potent inducer of autophagic cell death [95]. Remarkably, [PIC]PEI induced recurrent cycles of endosome and lysosome generation and degradation, which may reduce the threshold for cell viability and sensitize the cells to a parallel apoptotic pathway. Importantly, when autophagy was inhibited via Bafilomycin A, CQ, or atg5 deletion, cell death was suppressed. Furthermore, cell death was also reduced in melanoma cells upon pharmacological caspase inhibition or upon genetic deletion of Bax and Bak, two multi-domain Bcl-2 family members required for apoptosis; however, such manipulations had no effect on autophagy induction. Based on these results and the observed time lag between autophagy induction (2–5 h post-treatment) and final cell collapse (at 24–48 h), the authors speculated that early and persistent autophagy induction is required for apoptosis activation at a subsequent stage. Overall, these experiments illustrate the complex interplay between autophagy and apoptosis and corroborate that both autophagy and apoptosis are needed to kill cells in certain therapeutic contexts; in such situations, stimulating autophagy may enhance drug cytotoxicity.
Detecting autophagy in human cancer tissues
Despite these exciting experimental findings in cell-based and mouse models, it is important to recognize that no direct evidence exists showing that reduced or defective autophagy is a common requirement for human cancer initiation. This is primarily because the direct and quantitative detection of autophagy levels in human tumor samples remains technically intractable. As the importance of autophagy in carcinogenesis is unveiled, there is a pressing need for better detection methods and the identification of suitable markers of autophagy in human pathology samples [96]. Electron microscopy is still the most sensitive standard method for this purpose. The detection of autophagosomes (with intact cytoplasmic materials and organelles) and autolysosomes (with partially degraded materials) can be used to identify the different stages of autophagy by electron microscopy [97]. However, subclassification of these autophagic ultrastructures is extremely subjective (e.g., distinction of autolysosomes from other membrane-enclosed cellular compartments) and thus requires extensive expertise [98]. Moreover, due to the elaborate tissue processing steps involved, electron microscopy is labor-intensive and expensive, and its application is particularly unfeasible in clinical samples.
Therefore, even though electron microscopy remains the gold standard method for monitoring steady-state levels of autophagy, use of cell biological and biochemical methods are more practical approaches in detection and better quantification of autophagy and autophagic flux. Weakly basic dyes such as monodansylcadaverine and acridine orange were initially proposed as specific in vivo marker of autophagic vacuoles [99]. However, these dyes were later shown to have high affinity for lysosomes [100], and thus, their use is no longer recommended. As the molecular mechanism of autophagy is unraveling, better assays are being developed for reliable quantitative measurement of autophagy.
The biggest obstacle in developing methods to detect bona fide autophagy in clinical material arises from the fact that autophagy is a multi-step process characterized by the formation of an autophagosome, followed by its fusion to the lysosome and degradation of the autophagy substrates by lysosomal enzymes. As a result, an apparent increase in the number of autophagosomes observed within a cell can either mean an increase in the rate of autophagosome formation or alternatively, a decrease in the rate of autolysosome formation. Thus, the measurement of a dynamic process using static methods in clinical samples poses a fundamental challenge in data interpretation.
LC3 (ATG8)
The detection of LC3-II levels by fluorescence microscopy, immunoblotting, or immunohistochemistry is the most popular method for analysis of autophagy in cells. As discussed above, LC3-II is the lipidated form of LC3 resulting from conjugation to phosphotidylethanolamine by the ubiquitin-like conjugation system. LC3-II is incorporated onto both the outer and inner surfaces of the expanding autophagosome. After fusion with the lysosome, resident enzymes digest LC3-II associated with the inner surface, while LC3-II on the outer surface is degraded by ATG4. Since LC3 remains attached to the autophagosome until its final fusion with the lysosome, it serves as a useful and specific marker for the initial steps of autophagy. Although LC3 has three isoforms (LC3A, LC3B, and LC3C), LC3B levels correlates best with autophagic status1, and therefore, antibodies against this isoform are primarily used for the detection of autophagy.
GFP-LC3-overexpressing cells and transgenic animals are popular experimental models in which the transition from diffuse GFP-LC3 cytoplasmic staining to punctate localization in autophagosomes is used as readout of autophagy. Though GFP-LC3 puncta is ideally equivalent to autophagosome number, GFP-LC3 is prone to aggregate formation. Because these aggregates are difficult to distinguish from GFP-LC3 puncta corresponding to bona fide autophagosomes, the use of these fluorescent strategies can give rise to false-positive results [101]. Thus, assessing endogenous LC3-I to LC3-II conversion via immunoblotting is another useful tool to detect autophagy induction; LC3-II results in a faster migrating band due to high hydrophobicity of the conjugated PE. Furthermore, the determination of LC3-II levels in the presence and absence of chemical inhibitors blocking various steps of autophagy, (e.g., 3-methyladenine for inhibition of early autophagosome formation; bafilomycin, chloroquine, or E64D plus pepstatin A for inhibition of lysosomal activity) helps to dissect out the status of autophagic flux [102].
Moreover, with the emergence of more sensitive antibodies (Novus NBP1-19 167 against LC3B-II), LC3 immunohistochemical staining is a valuable tool to detect autophagosome levels in human tissue samples. However, caution should be exercised while relying on LC3 as the sole marker of autophagy since in certain cases, for e.g., in ATG14- or FIP200-deficient mouse fibroblasts, which do not show any autophagic activity, LC3 II levels are detected, indicating that LC3 can also be regulated in an autophagy-independent manner [96]. More importantly, the detection of punctate LC3 in tissues only provides information regarding autophagosome number rather than a readout of autophagic flux. Whereas lysosomal inhibition can be tractably employed in experimental systems for the determination of autophagic flux, such manipulations cannot be applied to clinical samples obtained from patients.
p62/SQSTM
Because of the limitations of using LC3 as an immunohistochemical marker to detect autophagy, there is a pressing need for alternative markers and techniques to more reliably measure autophagy in clinical samples. One attractive candidate is p62/SQSTM, a scaffold protein that interacts with polyubiquitinated protein aggregates and targets them to autophagosomes via its interaction with LC3 [103]. Importantly, p62 protein is degraded in an autophagy-dependent manner and its expression is inversely correlated with autophagic activity [96, 104]. Recently, it was shown that elimination of p62 via autophagy impedes tumorigenesis [66]. Thus, quantifying p62 levels in tumor tissues may be a useful marker for autophagy as well as tumorigenicity. However, because p62 is also regulated at the transcriptional level, its protein expression can be regulated independent of autophagy [105].
Overall, although the existing markers to detect and measure autophagy have proven effective in cell culture systems, it remains uncertain if such reagents and technique are suitable or adequate for studies of primary histopathology material. Thus, the discovery of additional markers that can be universally applied for autophagy detection is critical to utilize autophagy as a potential biomarker as well as to exploit this pathway therapeutically in cancer.
Concluding remarks
In summary, autophagy has emerged as an important regulator of tumorigenesis. In light of current evidence, it is hypothesized that autophagy maintains genomic integrity and thus prevents tumor initiation. On the other hand, in established tumors, the stress adaptive property of autophagy improves the cellular fitness of cancer cells and promotes their survival during disease progression and during anti-neoplastic therapy. In spite of these current dogmas, direct evidence establishing this stage-specific role of autophagy in carcinogenesis is missing. Thus, the development of mouse cancer models in which autophagy can precisely be manipulated is imperative to reveal important answers not only about the precise role of autophagy in different stages of carcinogenesis but also how autophagy levels can be regulated for optimal therapeutic benefits. Finally, there remains a need to identify robust tissue biomarkers for autophagy in human cancer specimens.
References
Suzuki K, Ohsumi Y (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 581:2156–2161
Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721
Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB (2005) Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120:237–248
Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169:425–434
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036
Debnath J, Baehrecke EH, Kroemer G (2005) Does autophagy contribute to cell death? Autophagy 1:66–74
Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132:27–42
Kondo Y, Kanzawa T, Sawaya R, Kondo S (2005) The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 5:726–734
Levine B (2007) Cell biology: autophagy and cancer. Nature 446:745–747
Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284:12297–12305
Hara T, Mizushima N (2009) Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy 5:85–87
Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S (2002) Truncating mutations of RB1CC1 in human breast cancer. Nat Genet 31:285–288
Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003
Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB (2008) Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112:1493–1502
Simonsen A, Tooze SA (2009) Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186:773–782
Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8:688–699
Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19:5360–5372
Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 11:468–476
Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11:385–396
Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447:1121–1125
Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2:211–216
Ohsumi Y, Mizushima N (2004) Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol 15:231–236
Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 29(11):1792–1802
Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19:2092–2100
Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282:37298–37302
Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, Kominami E, Tanaka K, Komatsu M (2008) The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 19:4762–4775
Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ (2007) Potential therapeutic applications of autophagy. Nat Rev Drug Discov 6:304–312
Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10:776–787
Chen N, Debnath J (2010) Autophagy and tumorigenesis. FEBS Lett 584:1427–1435
Efeyan A, Sabatini DM (2009) mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22:169–176
Neufeld TP (2010) TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 22:157–168
Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (2003) Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase—activating protein complex toward Rheb. Curr Biol 13:1259–1268
Manning BD, Cantley LC (2003) Rheb fills a GAP between TSC and TOR. Trends Biochem Sci 28:573–576
Bunney TD, Katan M (2010) Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer 10:342–352
Huang J, Manning BD (2009) A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans 37:217–222
Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, Davis DP, Stern HM, Murray LJ, Hoeflich KP, Klumperman J, Friedman LS, Lin K (2008) Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 183:101–116
Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E (2001) The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 276:35243–35246
Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL (2004) Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz–Jeghers syndrome. Genes Dev 18:1533–1538
Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30:214–226
Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO (2009) Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136:521–534
Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr (2004) Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18:2893–2904
Feng Z, Zhang H, Levine AJ, Jin S (2005) The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA 102:8204–8209
Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ (2007) The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 67:3043–3053
Budanov AV, Karin M (2008) p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134:451–460
Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G (2009) Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle 8:1571–1576
Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM (2006) DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126:121–134
Ciuffreda L, Di Sanza C, Milella M (2010) The mTOR Pathway: a new target in cancer therapy. Curr Cancer Drug Targets 10(5):484–495
Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, Fan J (2008) Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res 68:9167–9175
Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, Kinoshita T, Ueno T, Esumi H, Ochiai A (2008) Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci 99:1813–1819
Pirtoli L, Cevenini G, Tini P, Vannini M, Oliveri G, Marsili S, Mourmouras V, Rubino G, Miracco C (2009) The prognostic role of Beclin 1 protein expression in high-grade gliomas. Autophagy 5:930–936
Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676
Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T (2004) Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene 23:3898–3904
Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME (2009) ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem 284:2803–2810
Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, Kimchi A (2006) A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell 22:463–475
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939
Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T (2001) Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2:330–335
Yue Z, Jin S, Yang C, Levine AJ, Heintz N (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 100:15077–15082
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112:1809–1820
Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ (2008) Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol 39:1059–1063
Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, Pledger WJ, Wang H-G (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 9:1142–1151
Lee JW, Jeong EG, Soung YH, Nam SW, Lee JY, Yoo NJ, Lee SH (2006) Decreased expression of tumour suppressor Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric carcinomas. Pathology 38:312–315
Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C (2007) Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem 282:18573–18583
Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH (2009) Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol 217:702–706
Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E (2007) Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 21:1367–1381
Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21:1621–1635
Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137:1062–1075
Meek DW (2009) Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 9:714–723
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10:51–64
DeNardo DG, Johansson M, Coussens LM (2008) Immune cells as mediators of solid tumor metastasis. Cancer Metastasis Rev 27:11–18
Ostrand-Rosenberg S (2008) Immune surveillance: a balance between protumor and antitumor immunity. Curr Opin Genet Dev 8:11–18
Bingle L, Brown NJ, Lewis CE (2002) The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol 196:254–265
Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS (2008) Oncogene—induced senescence relayed by an interleukin—dependent inflammatory network. Cell 133:1019–1031
Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM, Narita M (2009) Autophagy mediates the mitotic senescence transition. Genes Dev 23:798–803
Bertout JA, Patel SA, Simon MC (2008) The impact of O2 availability on human cancer. Nat Rev Cancer 8:967–975
Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283:10892–10903
Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29:2570–2581
Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF (2007) BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol 27:6229–6242
Papandreou I, Lim AL, Laderoute K, Denko NC (2008) Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ 15:1572–1581
Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG (2010) The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 120:127–141
Fung C, Lock R, Gao S, Salas E, Debnath J (2008) Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 19:797–806
Aguirre-Ghiso JA (2007) Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 7:834–846
Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, Liao WS, Bast RC Jr (2008) The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest 118:3917–3929
Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB (2007) The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 9:218–224
Korah R, Boots M, Wieder R (2004) Integrin alpha5beta1 promotes survival of growth-arrested breast cancer cells: an in vitro paradigm for breast cancer dormancy in bone marrow. Cancer Res 64:4514–4522
White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ (2004) Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell 6:159–170
Livesey KM, Tang D, Zeh HJ, Lotze MT (2009) Autophagy inhibition in combination cancer treatment. Curr Opin Investig Drugs 10:1269–1279
Geser A, Brubaker G, Draper CC (1989) Effect of a malaria suppression program on the incidence of African Burkitt's lymphoma. Am J Epidemiol 129:740–752
Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB (2007) Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117:326–336
Maclean KH, Dorsey FC, Cleveland JL, Kastan MB (2008) Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest 118:79–88
Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, Galavotti S, Young KW, Selmi T, Yacobi R, Van Etten RA, Donato N, Hunter A, Dinsdale D, Tirro E, Vigneri P, Nicotera P, Dyer MJ, Holyoake T, Salomoni P, Calabretta B (2009) Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest 119:1109–1123
Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL (2007) Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 110:313–322
Kroemer G, Levine B (2008) Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 9:1004–1010
Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, Gonzalez-Feria L, Iovanna JL, Guzman M, Boya P, Velasco G (2009) Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest 119:1359–1372
Pardo R, Lo Re A, Archange C, Ropolo A, Papademetrio DL, Gonzalez CD, Alvarez EM, Iovanna JL, Vaccaro MI (2010) Gemcitabine induces the VMP1-mediated autophagy pathway to promote apoptotic death in human pancreatic cancer cells. Pancreatology 10:19–26
Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, Mulero F, Piris MA, Dash R, Barral PM, Rodriguez-Peralto JL, Ortiz-Romero P, Tuting T, Fisher PB, Soengas MS (2009) Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell 16:103–114
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140:313–326
Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2009) Monitoring autophagy by electron microscopy in Mammalian cells. Meth Enzymol 452:143–164
Eskelinen EL (2008) To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy 4:257–260
Biederbick A, Kern HF, Elsasser HP (1995) Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol 66:3–14
Mizushima N (2004) Methods for monitoring autophagy. Int J Biochem Cell Biol 36:2491–2502
Kuma A, Matsui M, Mizushima N (2007) LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3:323–328
Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3:542–545
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145
Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163
Moscat J, Diaz-Meco MT (2009) p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137:1001–1004
Shingu T, Fujiwara K, Bogler O, Akiyama Y, Moritake K, Shinojima N, Tamada Y, Yokoyama T, Kondo S (2009) Stage-specific effect of inhibition of autophagy on chemotherapy-induced cytotoxicity. Autophagy 5:537–539
Walker T, Mitchell C, Park MA, Yacoub A, Graf M, Rahmani M, Houghton PJ, Voelkel-Johnson C, Grant S, Dent P (2009) Sorafenib and vorinostat kill colon cancer cells by CD95-dependent and -independent mechanisms. Mol Pharmacol 76:342–355
Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA (2009) Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS ONE 4:e6251
Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM (2008) Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat 112:389–403
Ben Sahra I, Laurent K, Giuliano S, Larbret F, Ponzio G, Gounon P, Le Marchand-Brustel Y, Giorgetti-Peraldi S, Cormont M, Bertolotto C, Deckert M, Auberger P, Tanti JF, Bost F (2010) Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res 70:2465–2475
Kim RH, Coates JM, Bowles TL, McNerney GP, Sutcliffe J, Jung JU, Gandour-Edwards R, Chuang FY, Bold RJ, Kung HJ (2009) Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res 69:700–708
Shanmugam M, McBrayer SK, Qian J, Raikoff K, Avram MJ, Singhal S, Gandhi V, Schumacker PT, Krett NL, Rosen ST (2009) Targeting glucose consumption and autophagy in myeloma with the novel nucleoside analogue 8-aminoadenosine. J Biol Chem 284:26816–26830
Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C (2009) The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer 125:717–722
Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y (2005) Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol 26:1401–1410
Katayama M, Kawaguchi T, Berger MS, Pieper RO (2007) DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ 14:548–558
Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A (2007) Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 14:500–510
Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ (2008) A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell 14:90–102
Cao C, Subhawong T, Albert JM, Kim KW, Geng L, Sekhar KR, Gi YJ, Lu B (2006) Inhibition of mammalian target of rapamycin or apoptotic pathway induces autophagy and radiosensitizes PTEN null prostate cancer cells. Cancer Res 66:10040–10047
Acknowledgements
JD is supported by grants from the NIH (RO1CA126792; CA126792-S1 ARRA), the California Tobacco Related Disease Research Program (18XT-0106), and an HHMI Physician Scientist Early Career Award.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is published as part of the Special Issue on Autophagy.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Roy, S., Debnath, J. Autophagy and Tumorigenesis. Semin Immunopathol 32, 383–396 (2010). https://doi.org/10.1007/s00281-010-0213-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-010-0213-0