Abstract
Diffuse Large B Cell Lymphoma (DLBCL) is the most common form of blood cancer. Among the subtypes, the activated B-cell (ABC) subtype is typically more aggressive and associated with worse outcomes. However, the underlying mechanisms are not fully understood. In this study, we performed microarray analysis to identify potential ABC-DLBCL-associated genes. We employed Kaplan–Meier methods and cox univariate analysis to explore the prognostic value of the identified candidate gene Coiled-coil domain containing 50 (CCDC50). Additionally, we used DLBCL cell lines and mouse models to explore the functions and mechanisms of CCDC50. Finally, we isolated CCDC50-bearing exosomes from clinical patients to study the correlation between these exosomes and disease severity. Our results demonstrated that CCDC50 not only showed significantly positive correlations with ABC subtype, tumor stage and number of extranodal sites, but also suggested poor outcomes in DLBCL patients. We further found that CCDC50 promoted ABC-DLBCL proliferation in vitro and in vivo. Mechanistically, CCDC50 inhibited ubiquitination-mediated c-Myc degradation by stimulating the PI3K/AKT/GSK-3β pathway. Moreover, CCDC50 expression was positively correlated with c-Myc at protein levels in DLBCL patients. Additionally, in two clinical cohorts, the plasma CCDC50-positive exosomes differentiated DLBCL subtypes robustly (AUC > 0.80) and predicted disease severity effectively (p < 0.05). Our findings suggest that CCDC50 likely drives disease progression in ABC-DLBCL patients, and the CCDC50-bearing exosome holds great potential as a non-invasive biomarker for subtype diagnosis and prognosis prediction of DLBCL patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diffuse Large B Cell Lymphoma (DLBCL), the most common type of non-Hodgkin lymphoma (NHL), account for 30 ~ 40% of all newly diagnosed cases worldwide [1]. Despite the standard frontline chemoimmunotherapy regimen (R-CHOP) improving the outcomes of DLBCL patients, up to 40% of patients still succumb to this disease. Immunohistochemistry analysis categorizes DLBCL into GCB and non-GCB subtypes based on the expression levels of MUM1, BCL6, and CD10 [2]. Non-GCB subtype patients have a shorter survival time. Gene expression profiling has identified two major subtypes: germinal center B cell-like (GCB) and activated B cell-like (ABC) DLBCL [3]. ABC-DLBCL is more chemo-resistant and has an inferior prognosis compared to the GCB subtype, with a 3-year progression-free survival rate of 40% versus 75% (p < 0.001) [1, 4,5,6]. Despite significant advances over the past few decades, oncogenic aberrations in ABC-DLBCL progression are not fully characterized. Thus, the ongoing effort is required to identify new biomarkers associated with this aggressive subtype.
Currently, the diagnosis of DLBCL subtypes primarily relies on invasive procedures, such as fine needle aspiration, core biopsy, and lymph node excision. These methods present challenges in obtaining a quick and accurate diagnosis. Thus, the identification of sensitive biomarkers with non-invasive ways, such as reliable liquid biopsy, is highly desired. Numerous studies have shown that tumor derived exosomes can be effective biomarkers for cancer diagnosis [7]. However, their potential to diagnose and predict the progression of DLBCL remains poorly defined.
CCDC50 is a tyrosine-phosphorylated protein with multiple ubiquitin-interacting domains [8]. Recent studies have shown that CCDC50 acts as a new autophagy receptor, suppressing antiviral signaling by negatively regulating RIG-I/MADA5 [9], cGAS-STING [10], and NLRP3 [11]. In Hepatocellular Carcinoma (HCC) [12], Mantle Cell Lymphoma (MCL) and Chronic Lymphocytic Leukemia (CLL) [13], CCDC50 is required for cell survival. However, its function and molecular consequences in DLBCL remain unclear to date.
In this study, we present the following discoveries: (1) CCDC50 is increased in ABC-DLBCL and associated with poor prognosis, (2) CCDC50 stabilizes c-Myc protein in a PI3K/AKT/GSK-3β dependent manner, leading to the proliferation of ABC-DLBCL, and (3) DLBCL-derived CCDC50-positive exosomes in plasma effectively distinguish DLBCL subtypes and predictes patient severity. These findings may help develop a new therapeutic strategy to treat ABC-DLBCL and provide a non-invasive approach for subtype diagnosis and prognostic monitoring of DLBCL.
Materials and methods
Data collection and bioinformatic analysis
GSE10846, GSE19246, GSE32918, GSE50721, GSE64820, and GSE94669, were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). DLBCL datasets from the GEPIA (Gene Expression Profiling Interactive Analysis) (http://gepia.cancer-pku.cn) and the TCGA (The Cancer Genome Atlas) (https://xenabrowser.net/datapages/) were also used in this study. A total of 284 paraffin-embedded tissue samples and 129 plasma samples of DLBCL patients were collected from The First Affiliated Hospital, Zhejiang University School of Medicine.
Limma was used to identify differentially expressed genes, and two R packages (survival, survminer) were used to plot Kaplan–Meier curves. GSE10846 and 65 paraffin-embedded tissue samples with cilinical information were used for survival analysis.
Patients were divided into the high and low expressed groups according to the transcriptional value of CCDC50 (high: value > upper quartiles, low: value < lower quartiles). GSEA software (version 4.0.3) was used to identify tumor-related pathways in the CCDC50 high and low groups.
Cell culture, plasmids, transfection
Human DLBCL cell lines (SU-DHL-2, OCI-LY3, OCI-LY10, SU-DHL-4, Pfeiffer, and Toledo) were grown in IMDM with 10% FBS. HEK293T and Hela cells were grown in DMEM with 10% FBS. SU-DHL-2, Pfeiffer, and Toledo were purchased from Shanghai EK-Bioscience Co., Ltd; SU-DHL4 was obtained from the Type Culture Collection of the Chinese Academy of Sciences; OCI-Ly3 and OCI-LY10 were kindly provided by Dr. Zhe Wang from the Department of Pathology at Xijing Hospital, the Fourth Military Medical University. HEK293T and Hela were kindly provided by Dr. Chengfang Dong from Department of Pathology, Zhejiang University School of Medicine. All the cells were grown in a humidified atmosphere with 5% CO2 at 37 °C.
The shRNA sequence was cloned into pLVX-shRNA1. The sequence of c-Myc (NM_ 001354870.1) and CCDC50 (NM_174908.4) were cloned into Plvx-IRES-Neo and pLVX-TRE3G, respectively. Stable transfection cells were selected with puromycin (2 μg/ml) and/or G418 (400 μg/ml). Lipo3000 Transfection Reagent (Invitrogen) was used for lentivirus production. Sequence information of shRNA is summarized in Table S1.
Real-time qPCR, Western blotting (WB), CCK8, EdU assays, Immunoprecipitation, immunofluorescence (IF), Immunohistochemistry (IHC), and antibodies
Experiments were performed as described previously [14]. In our Real-time qPCR experiments, the primer sequences employed are summarized in Table S1. The primer for the c-Myc was adopted from a previously published literature [15]. The primer of CCDC50 was designed according to the sequence of CCDC50 variant 1. We then performed PrimerBLAST analysis to ensure its accurateness and specificity. The concentration of antibodies used in IF: c-Myc (1:100, ab32072). The concentration of antibodies used in IHC: CCDC50 (1:500, ab127169), c-Myc (1:100, ab32072). The Staining value (values 0–12) in IHC experiments was calculated by the product of the intensity of CCDC50 positive staining (negative, 0; weak 1; moderate, 2; strong, 3 scores) and the proportion of immunopositive cells of interest (< 25%, 1; 25–50%, 2; 50–75%, 3; > 75%, 4 scores). Patients were divided into two groups according to median value of IHC staining value. Primary and secondary antibodies used in this study were listed in Table S2.
Xenograft studies in NOD/SCID mice
5 × 106 cells were collected and mixed with 50uL PBS and 50uL Matrigel (Corning 354234). Cells transfected with shNTC were injected into the right flank of 6–8 weeks old NOD-SCID mice, and cells transfected with shCCDC50 were injected into the left flank. The tumor volumes were measured every 5 days after 5 days post engraftment.
Inhibitor and ubiquitination assay
Cycloheximide (CHX, 100ug/ml, MCE) was used to inhibit protein synthesis. MG132 (10 μM, MCE) was used to inhibit proteasome proteolytic activity. CHIR-99021 (1 μM, MCE) was used to inhibit GSK-3β activity.
1 × 106 cells were seeded per well in 6-well plates and transfected with indicated plasmids using PolyJetTM DNA In Vitro Transfection Reagent (SigaGen). After a 48-h incubation period, 10uM of MG132 was added to the culture medium, and cells were incubated for an additional 6 h. Then, cells were lysed in cell lysis buffer (Beyotime, P0013) and subjected to immunoprecipitation using an anti-c-Myc antibody (Abcam, ab32072). The anti-K48-Ub antibody (CST #4289) was used to detect the ubiquitination levels of c-Myc.
Exosome extraction, nanoparticle tracking analysis, and Apogee nanoscale flow cytometry analysis
The protocols of these experiments were detailed in our recent study [16]. We also summarized the exosome extraction and analysis in Supplementary file 1. Specifically, rabbit anti-CCDC50 monoclonal antibodies were labeled with Zenon Alexa Fluor 647. Rabbit anti-IgG monoclonal antibodies were also labeled as negative controls.
Electron microscopy
10 μL of exosome liquid was deposited onto a 300-mesh carbon-coated copper grid and incubated for 1 min. Subsequently, the exosome liquid was carefully removed using filter papers. Then, 10 μL of a 2% uranyl acetate staining solution was deposited to the copper grid and incubated for 1 min, followed by removal of the solution using filter paper. Next, 10 μL of staining solution was deposited to the copper grid and incubated for 1 min, after which the staining solution was removed using filter paper. Finally, the morphology of the exosomes was observed using a transmission electron microscope (Talos 120 kV cryo-TEM).
Statistical analysis
All statistical analyses including unpaired t-test, one-way ANOVA, Pearson and Spearman correlation analyses, and Chi-Square Test were performed using GraphPad Prism9 software (version 9.2.0). Experiments were repeated independently no less than three times with results presented as the mean ± SD. P < 0.05 was considered statistically significant. The following symbols were used to denote the level of significance: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
CCDC50 is upregulated in ABC-DLBCL and associated with inferior prognosis
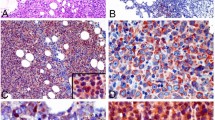
We conducted bioinformatic and survival analyses to identify driver oncogenes associated with ABC-DLBCL (Fig. S1). In this study, we found that CCDC50 effectively discriminate ABC from GCB-DLBCL with AUC values ranging from 0.7871 to 1.0000 (Fig. 1A-B). Analyses of two independent datasets showed that CCDC50 mRNA levels were significantly elevated in DLBCL patients compared to healthy controls (Fig. 1C-D). Similarly, CCDC50 protein levels were higher in DLBCL than in lymphoid tissues (Fig. 1E). Next, we investigated the relationships between CCDC50 and clinicopathologic features in patients (GSE10846, Table S3). We observed significant correlations between CCDC50 expression and DLBCL subtype, tumor stage and number of extranodal sites (p < 0.05, Table 1, n = 414). Kaplan–Meier survival analysis revealed that patients with higher CCDC50 expression showed shorter overall survival (p = 0.031, Fig. 1F, Supplementary file 2) and disease free survival time (p = 0.026, Fig. 1G). Furthermore, the results of cox univariate analysis corroborated these findings, clearly identifying CCDC50 as a significantly risk factor for DLBCL patients (Fig. 1H). Our IHC experiment also showed that higher CCDC50 expression is significantly associated with shorter overall survival time in DLBCL (p < 0.0001, Fig. 1I, Supplementary file 3). Collectively, our results suggest that CCDC50 is significantly elevated in DLBCL, especially in ABC-DLBCL, and is a predictor of worse prognosis.
CCDC50 is upregulated in ABC-DLBCL and associated with poor prognosis. Dot plots indicated CCDC50 mRNA expression in DLBCL subtypes and ROC curves indicated the accuracy of DLBCL subtype diagnosis in cell line datasets (A), and patient datasets (B). CCDC50 mRNA (C, D) and protein levels (E) in DLBCL and normal samples. Kaplan–Meier survival analysis (F, G) and cox univariate analysis (H) of CCDC50 mRNA expression in DLBCL datasets. (I) Kaplan–Meier survival analysis of CCDC50 protein expression in DLBCL patients
CCDC50 promotes ABC-DLBCL proliferation in vitro and in vivo
We performed RT-qPCR and WB analysis to assess CCDC50 expression in DLBCL cell lines. Results revealed that both CCDC50 mRNA and protein levels were significantly higher in ABC-DLBCL cell lines (Fig. 2A). We then selected ABC-DLBCL cell lines for further study. We knocked down CCDC50 in OCI-LY3/SU-DHL-2 and overexpressed it in OCI-LY10 (Fig. 2B). Results of CCK8 and EdU assays showed that CCDC50 knockdown significantly reduced cell proliferation, while CCDC50 overexpression remarkably increased it (Fig. 2C, Supplementary file 4, Fig. 2D). Furthermore, we established a xenograft model by subcutaneously injecting CCDC50 stably knocked down OCI-LY3/SU-DHL-2 cells. Tumors in shCCDC50 group displayed slower growth than those in shNTC group (Fig. 2E), ultimately reducing the tumor mass (Fig. 2F). Additionally, tumors in the shCCDC50 group exhibited decreased Ki67 staining values (Fig. 2G). These results confirm that CCDC50 is essential for ABC-DLBCL proliferation both in vitro and in vivo.
CCDC50 promotes ABC DLBCL proliferation in vitro and in vivo. (A) CCDC50 expression in DLBCL cell lines. (B) CCDC50 mRNA and protein levels in shNTC-/shCCDC50-OCI-LY3 cells, shNTC-/shCCDC50-SU-DHL-2 cells, and vector/CCDC50-OCI-LY10 cells. CCK8 (C) and EdU assays (D) were performed to detect the proliferation of shNTC-/shCCDC50-OCI-LY3 cells, shNTC-/shCCDC50-SU-DHL-2 cells, and vector/CCDC50-OCI-LY10 cells. (E) The tumor growth curve. (F) tumor weight. (G) Tumor tissue Ki67 IHC staining in NOD-SCID mice
c-Myc is the downstream effector of CCDC50-induced ABC-DLBCL proliferation
We performed GSEA analysis to uncover molecular mechanisms behind CCDC50-induced proliferation. Results revealed that patients with higher expression of CCDC50 exhibited significant enrichment of MYC-related pathways (Fig. 3A). Moreover, patients in the TCGA dataset with higher expression of CCDC50 showed elevated levels of c-Myc-induced genes (Fig. 3B). We hypothesized that CCDC50 promotes ABC-DLBCL proliferation via c-Myc. Immunofluorescence experiments indicated that after the knockdown of CCDC50, the levels of c-Myc decreased, whereas overexpression of CCDC50 resulted in a notable accumulation of c-Myc (Fig. 3C), which was supported by WB analysis (Fig. 3D). To confirm that c-Myc is the downstream target of CCDC50-induced ABC-DLBCL proliferation, we enforced the expression of c-Myc in cells with CCDC50 knockdown (Fig. 3E). Results of CCK8 and EdU assays showed that c-Myc recovery rescued cell proliferation arrest caused by CCDC50 knockdown (Fig. 3F-G). These findings suggest that CCDC50 promotes tumor proliferation by positively regulating c-Myc expression in ABC-DLBCL.
CCDC50 promotes ABC-DLBCL proliferation through positively regulate c-Myc. (A) Enriched pathways were detected by the GSEA approach based on patient datasets. (B) The heat map represented different expressions of c-Myc target genes in CCDC50-high and CCDC50-low groups in TCGA dataset. (C, D) c-Myc expression in shNTC-/shCCDC50-OCI-LY3, shNTC-/shCCDC50-SU-DHL-2, and vector/CCDC50-OCI-LY10 cells. (E) The stable clone with c-Myc expression was established in CCDC50-depleted cells. (F, G) The cell viability states and percentage of proliferated cells before and after overexpression of c-Myc in CCDC50-depleted cells
CCDC50 stabilizes c-Myc protein by reducing its ubiquitination
We next explore whether CCDC50 affects c-Myc expression at the transcriptional or post-transcriptional level. We found that knockdown of CCDC50 increased c-Myc mRNA expression in OCI-LY3, while overexpression of CCDC50 decreased c-Myc mRNA expression in OCI-LY10 (Fig. 4A). Meanwhile, no significant correlations were detected between CCDC50 and c-Myc mRNA (Fig. 4B), indicating that CCDC50 primarily regulates c-Myc post-translationally. Through CHX chase experiments, we observed that CCDC50 knockdown decreased the half-life of the endogenous c-Myc protein, while CCDC50 overexpression increased it (Fig. 4C). Furthermore, treatment with MG132 restored c-Myc protein levels in CCDC50-knocked down cells (Fig. 4D), indicating that the regulation may be involved in ubiquitination process. We then examined the phosphorylation of c-Myc at the threonine 58, which results in ubiquitination-mediated degradation of c-Myc. Results showed that CCDC50 knockdown increased the p-T58 level, while CCDC50 overexpression decreased it (Fig. 4E). Moreover, CCDC50 knockdown significantly increased c-Myc ubiquitination, while CCDC50 overexpression dramatically repressed it (Fig. 4F). These findings support that CCDC50 stabilizes c-Myc by preventing its ubiquitination.
CCDC50 stabilizes c-Myc protein by reducing its ubiquitination through PI3K/AKT/GSK-3β. (A) c-Myc mRNA level in cells with CCDC50 knockdown or CCDC50 expression. (B) Correlation between c-Myc and CCDC50 at mRNA levels in DLBCL. (C) Cells were treated with CHX for indicated time intervals and the changes of c-Myc expression. (D) Cells with CCDC50 knockdown were treated with MG-132 for 4 h and the changes of c-Myc expression were examined. (E) The phosphorylated level of c-MycT58 in cells with CCDC50 knockdown or overexpression. (F) Ubiquitination and phosphorylation levels of c-Myc in shNTC-/shCCDC50-Hela cells and vector-/CCDC50-HEK-293 T cells. (G) Enriched pathways in CCDC50-high groups in DLBCL patient. (H) The protein level of PI3K/AKT/GSK-3β pathways, and CCDC50 in shNTC-/shCCDC50-OCI-LY3 cells, shNTC-/shCCDC50-SU-DHL-2 cells, and vector/CCDC50-OCI-LY10 cells. (I) Cells with CCDC50 knockdown were treated with CHIR-99021 (1 μM) for 4 h, and the protein level of c-Myc, p-GSK-3βY216 were examined
GSEA analysis revealed that high CCDC50 expression is associated with activated PI3K/AKT/mTOR pathway (Fig. 4G). Since active AKT inhibits the GSK-3β [17], preventing it from phosphorylating c-MycT58. CCDC50 may stabilize c-Myc protein via PI3K/AKT/GSK-3β pathway. As shown in Fig. 4H, CCDC50 knockdown reduced phosphorylation levels of p85 and AKT, while CCDC50 overexpression increased them. The level of active form p-GSK-3βY216 was increased after CCDC50 knockdown, but notably decreased after CCDC50 overexpression. The change of inactive form p-GSK-3βS9 showed the opposite trend. Additionally, treatment with CHIR-9902 significantly restored the level of c-Myc protein in cells with CCDC50 knockdown (Fig. 4I). These findings support that CCDC50 activates PI3K/AKT/GSK-3β axis to reduce ubiquitination-mediated c-Myc degradation.
CCDC50 expression is positively correlated with c-Myc in DLBCL
We then performed IHC staining to examine the correlation between CCDC50 and c-Myc. Patients with high CCDC50 expression exhibit high c-Myc expression (Fig. 5A). Approximately 56.32% of patients with higher CCDC50 expression display high c-Myc expression, whereas 43.68% showed weak c-Myc staining. Similarly, 70.87% of patients with low CCDC50 expression display weak c-Myc expression, whereas 29.13% showed high c-Myc staining (Fig. 5B). Above results indicated that the protein level of CCDC50 and c-Myc were positively correlated in DLBCL patients. Thus, CCDC50 could promote DLBCL proliferation in a PI3K/AKT/GSK-3β/c-Myc dependent manner (Fig. 5C).
CCDC50 expression is positively correlated with c-Myc. (A) c-Myc expression was higher in patients with higher expression of CCDC50. (B) Correlation between CCDC50 and c-Myc expression were assessed with Chi-Square Test. (C) A proposed model to illustrate that higher expression of CCDC50 activates AKT, which inhibits GSK-3β mediated phosphorylation of c-MycT58, therefore promoting c-Myc accumulation
Exosomal CCDC50 serves as a non-invasive diagnostic and prognostic biomarker in DLBCL
CCDC50 protein was found in exosomes released from DLBCL cell lines [18], prompting us to wonder whether plasma exosomes contain CCDC50 protein and whether it has clinical significance in DLBCL. We used Cryo-electron microscopy (Fig. 6A) and NTA scatter (Fig. 6B) to validate that we successfully isolated plasma exosomes from DLBCL patients. Then, based on flow cytometry analysis, we detected CCDC50 proteins in plasma exosomes (Fig. S2A). Because CD20 is a biomarker of DLBCL (Fig. S2B) and detected in plasma exosomes (Fig. S2C, Supplementary file 5), we regarded CD20 + exosomes as DLBCL-derived exosomes and CCDC50 + exosomes out of CD20 + exosomes as DLBCL-derived CCDC50 + exosomes. Results showed that the percentage of DLBCL-derived CCDC50 + exosomes were higher in Non-GCB compared to GCB in both cohorts (Fig. 6C, AUC = 0.9355; Fig. 6D, AUC = 0.8135). Moreover, based on the total IPI score, patients are divided into low- (0–1), intermediate- (2–3), and high-risk categories (4–5). We discovered that DLBCL-derived CCDC50 + exosomes were significantly higher in high-risk categories (Fig. 6E-F). Overall, the DLBCL-derived CCDC50 + exosome is a promising non-invasive biomarker with diagnostic and prognostic potential.
Plasma Exosomal CCDC50 protein serves as a non-invasive biomarker in DLBCL. The shape (A) and the diameter distribution (B) of plasma exosomes in DLBCL. The percentage of CCDC50 + exosomes out of CD20 + exosomes and ROC curves in the discovery (C) and validation cohort (D). The percentage of CCDC50 + exosomes out of CD20 + exosomes in different risk groups of DLBCL patients in the discovery (E) and validation cohort (F)
Discussion
DLBCL is a complex and heterogeneous disease. The ABC-DLBCL subtype is known for its aggressive nature and poor clinical outcomes. Despite extensive research, the underlying mechanisms involved in the pathogenesis of ABC-DLBCL remain poorly understood. To explore potential biomarkers and therapeutic targets, we conducted a transcriptome analysis and identified CCDC50, a poorly investigated gene that is dysregulated in DLBCL (Fig. 1).
The human CCDC50 gene has two isoforms: CCDC50 variant 1 (CCDC50-V1) contains 306 amino acids with skipping of exon 6, and CCDC50 variant 2 (CCDC50-V2) contains 482 amino acids. CCDC50-V1 is predominantly expressed and play oncogenic roles in HCC, whereas CCDC50-V2 is less expressed and has anti-tumor functions [12]. Similarly, in DLBCL, CCDC50-V1 is the dominant form, while CCDC50-V2 is rarely expressed (Fig. S3). Therefore, in this study, both knockdown and overexpression sequences of CCDC50 were targeted against CCDC50-V1. It has been reported that CCDC50 acts as a downstream effector of several receptors and functions as a negative regulator in NF-κB activity [19,20,21]. However, another study showed that CCDC50 maintains long-term activation of NF-κB signaling in Fas-stimulated thymocytes [22], and CCDC50 is required for tumor survival and activates NF-κB signaling in CLL and MCL [13]. The above data suggests that CCDC50 functions differently depending on the cellular context. In this study, we found that CCDC50 accelerates ABC-DLBCL proliferation both in vitro and in vivo (Fig. 2).
Subsequently, in exploring the biological mechanism of CCDC50 in ABC-DLBCL, we present multiple evidence supporting that CCDC50 could stabilize c-Myc to promote tumor proliferation (Fig. 3). The protein c-Myc plays critical roles in maintaining normal cell functions, including controlling cell growth and determining cell fate. The rapid degradation of c-Myc by the ubiquitin–proteasome system is essential for maintaining its normal physiological level [23, 24]. This degradation process involves sequential phosphorylation of c-Myc, starting with the stabilization of c-Myc upon the phosphorylation of S62 by EKR [25], followed by the GSK-3β mediated T58 phosphorylation [26], and finally recognition by E3 ligases [27, 28] and degradation by 26S proteasome. In many cancers, the molecules involved in c-Myc degradation pathways are dysregulated, leading to increased c-Myc stability. So far, direct targeting of c-Myc has not yet been successful due to its disordered structure and lack of specific enzymatic activity [29]. Hence, disrupting the molecules that regulate c-Myc degradation may have therapeutic benefits. In this study, we found that CCDC50 activates the PI3K/AKT pathway in DLBCL. Then, active AKT inhibits GSK3β function and reduces GSK3β-mediated phosphorylation of c-MycT58 (Fig. 4). Hence, our findings provide insight into the underlying mechanism of CCDC50-induced proliferation in ABC-DLBCL and further advance the understanding of the regulatory mechanisms that govern c-Myc stability.
Given the reliability and effectiveness of CCDC50 as a biomarker in DLBCL, we aspire to conduct further investigations to determine the feasibility of its application as a clinical biomarker.
Liquid biopsy is a minimally invasive detection method that analyzes various biomarkers in biofluids for disease diagnosis and prognosis monitoring [30]. Exosomes are important source of biomarkers in liquid biopsy as they carry multiple molecules of parent cells and are enriched in biofluids [7, 31, 32]. By analyzing the contents in exosomes, researchers can identify biomarkers for diseases. In DLBCL, several studies explored the clinical value of plasma exosomes. Feng Y found that exosomal miR-99a-5p and miR-125b-5p were effective in distinguishing chemosensitive and chemoresistant DLBCL, with AUC values of 0.744 and 0.7802, respectively, and both were associated with poor outcomes [33]. Cao D identified five exosomal miRNAs (miR‐379‐5p, miR‐135a‐3p, miR‐4476, miR‐483‐3p, and miR‐451a) that were differentially expressed in DLBCL patients and healthy individuals, with AUC values greater than 0.85 [34]. Carvalho reported that exosomes from ABC-DLBCL cell lines contained more CCDC50 proteins than GCB cell lines, indicating that DLBCL-derived exosomes carry CCDC50 proteins and exosomal CCDC50 showed significantly different levels in DLBCL subtypes [18]. Therefore, we questioned whether CCDC50 is present in the exosomes in DLBCL patients? If CCDC50 is present, whether these exosomes can serve as biomarkers when they enter the bloodstream? In this study, we discovered the presence of CCDC50 + exosomes derived from DLBCL in patients plasma. Further analysis revealed that these exosomes showed higher levels in Non-GCB DLBCL and higher risk patients (Fig. 6C-F). Taken together, these findings support that the DLBCL-derived CCDC50 + exosome could be a promising non-invasive biomarker for predicting patient outcomes and distinguishing between different subtypes of DLBCL. While our consideration of CD20 + exosomes as originating from DLBCL was supported by the significantly higher expression of CD20 in DLBCL compared to corresponding normal tissues (Fig. S2B), we acknowledge the potential for CD20 expression overlap on exosomes derived from both DLBCL and normal B-cells. As such, further research efforts should aim to identify a more specific biomarker for DLBCL, ultimately enhancing diagnostic accuracy in clinical settings.
Our work provides evidence that CCDC50 contributes to the proliferation of ABC-DLBCL by reducing ubiquitination-mediated c-Myc degradation. More importantly, our analysis of more than 1000 patient samples suggests that CCDC50 may serve as a promising diagnostic and prognostic biomarker in DLBCL. Therefore, the evaluation of CCDC50 as a treatment strategy for DLBCL patients warrants further clinical investigation.
Data availability
All the data corresponding to the DLBCL datasets used in this study are available in GEO, GEPIA, and TCGA database. All of the experiment data in this study are available with reasonable requested.
References
Lenz G, Staudt LM (2010) Aggressive lymphomas. N Engl J Med 362:1417–1429
Hans CP, Weisenburger DD, Greiner TC et al (2004) Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103:275–282
Alizadeh AA, Eisen MB, Davis RE et al (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503–511
Chapuy B, Stewart C, Dunford AJ et al (2018) Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24:679–690
Dunleavy K, Pittaluga S, Czuczman MS et al (2009) Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 113:6069–6076
Lenz G, Wright G, Dave SS et al (2008) Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 359:2313–2323
Yu D, Li Y, Wang M et al (2022) Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer 21:56–56
Vazza G, Picelli S, Bozzato A, Mostacciuolo ML (2003) Identification and characterization of C3orf6, a new conserved human gene mapping to chromosome 3q28. Gene 314:113–120
Hou P, Yang K, Jia P et al (2021) A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res 31:62–79
Hou P, Lin Y, Li Z et al (2021) Autophagy receptor CCDC50 tunes the STING-mediated interferon response in viral infections and autoimmune diseases. Cell Mol Immunol 18:2358–2371
Hou P, Tian T, Jia P et al (2023) The regulation of NLRP3 inflammasome activation by CCDC50-mediated autophagy. Autophagy 19(1):365–366
Wang H, Zhang CZ, Lu SX et al (2019) A coiled-coil domain containing 50 splice variant is modulated by Serine/Arginine-Rich splicing factor 3 and promotes hepatocellular carcinoma in mice by the ras signaling pathway. Hepatology 69:179–195
Farfsing A, Engel F, Seiffert M et al (2009) Gene knockdown studies revealed CCDC50 as a candidate gene in mantle cell lymphoma and chronic lymphocytic leukemia. Leukemia 23:2018–2026
Zhou L, Ding L, Gong Y et al (2021) NEK2 promotes cell proliferation and glycolysis by regulating PKM2 abundance via phosphorylation in diffuse large B-cell lymphoma. Front Oncol 11:677763
Garnar-Wortzel L, Bishop TR, Kitamura S et al (2021) Chemical inhibition of ENL/AF9 yeats domains in acute leukemia. ACS Cent Sci 7:815–830
Tian C, Stewart T, Hong Z et al (2023) Blood extracellular vesicles carrying synaptic function- and brain-related proteins as potential biomarkers for Alzheimer’s disease. Alzheimer’s Dement 19:909–923
Cross DA, Alessi DR, Cohen P et al (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785–789
Carvalho AS, Baeta H, Henriques AFA et al (2021) Proteomic landscape of extracellular vesicles for diffuse large B-cell lymphoma subtyping. Int J Mol Sci 22:11004
Zhi X, Chen Q, Song S et al (2020) Myostatin promotes osteoclastogenesis by regulating Ccdc50 Gene expression and RANKL-induced NF-κB and MAPK pathways. Front Pharmacol 11:565163
Bohgaki M, Tsukiyama T, Nakajima A et al (2008) Involvement of Ymer in suppression of NF-kappaB activation by regulated interaction with lysine-63-linked polyubiquitin chain. Biochem Biophys Acta 1783:826–837
Kameda H, Watanabe M, Bohgaki M et al (2009) Inhibition of NF-kappaB signaling via tyrosine phosphorylation of Ymer. Biochem Biophys Res Commun 378:744–749
Tsukiyama T, Matsuda-Tsukiyama M, Bohgaki M et al (2012) Ymer acts as a multifunctional regulator in nuclear factor-κB and Fas signaling pathways. Mol Med (Cambridge, Mass.) 18:587–597
Dang CV (2012) MYC on the path to cancer. Cell 149(1):22–35
Sears RC (2004) The life cycle of C-myc: from synthesis to degradation. Cell Cycle (Georgetown, Tex.) 3:1133–1137
Bachireddy P, Bendapudi PK, Felsher DW (2005) Getting at MYC through RAS. Clin Cancer Res 11:4278–4281
Gregory MA, Qi Y, Hann SR (2003) Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem 278:51606–51612
Yada M, Hatakeyama S, Kamura T et al (2004) Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23:2116–2125
Welcker M, Orian A, Jin J et al (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA 101:9085–9090
Massó-Vallés D, Soucek L (2020) Blocking myc to treat cancer: reflecting on two decades of omomyc. Cells 9(4):883
Ignatiadis M, Sledge GW, Jeffrey SS (2021) Liquid biopsy enters the clinic — implementation issues and future challenges. Nat Rev Clin Oncol 18:297–312
Zhou B, Xu K, Zheng X et al (2020) Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther 5:144
Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367(6478):eaau6977
Feng Y, Zhong M, Zeng S et al (2018) Exosome-derived miRNAs as predictive biomarkers for diffuse large B-cell lymphoma chemotherapy resistance. Epigenomics 11:35–51
Cao D, Cao X, Jiang Y et al (2022) Circulating exosomal microRNAs as diagnostic and prognostic biomarkers in patients with diffuse large B-cell lymphoma. Hematol Oncol 40:172–180
Funding
This study was supported by National Natural Science Foundation of China (81571226, 82020108012, and 81671187), Medical Science and Technology Program of Zhejiang Province (2018KY364), and Zhejiang Provincial Natural Science Foundation of China (No. LQ20H160025).
Author information
Authors and Affiliations
Contributions
Conceptualization, Yuqi Gong, Zhengrong Mao, Jing Zhang and Ren Zhou; Formal analysis, Yuqi Gong; Funding acquisition, Jing Zhao, Zhengrong Mao, Jing Zhang; Investigation, Yuqi Gong; Methodology, Fang Yu; Resources, Hongyan Tong, Fang Yu, Xianbo Huang, Guoping Ren, Zhongqin Fan and Zhe Wang; Supervision, Ren Zhou; Validation, Yuqi Gong, Qi Liu; Visualization, Yuqi Gong; Writing – original draft, Yuqi Gong; Writing – review & editing, Zhengrong Mao, Jing Zhang and Ren Zhou.
Corresponding authors
Ethics declarations
Ethics approval
This study was approved by the Ethics Committee of Zhejiang University.
Consent for publication
All authors signed informed consent regarding publishing the data.
Conflict of interest
All authors declare no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

277_2023_5409_Fig7_ESM.png
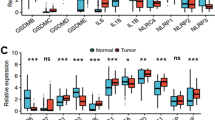
Fig. S1 The screening process of ABC-DLBCL-associated genes. (A) The work flow of the screening process. (B)The four genes in the left column were well investigated in ABC-DLBCL, the four genes in the middle-left column were reported in ABC-DLBCL, the nine genes in the middle-right column were mentioned in ABC-DLBCL, the thirteen genes in the right column never be reported in ABC-DLBCL. (PNG 482 kb)

277_2023_5409_Fig8_ESM.png
Fig. S2 Detection of Exosomal CD20 and CCDC50 proteins. (A) Levels of exosomal CD20 and CCDC50 proteins and their corresponding controls (IgG) were detected in DLBCL patient samples by flow cytometry. (B) The expression level of CD20 in 31 types of cancer. (C) The percentage of CD20+ exosomes out of all exosomes were detected in DLBCL patients and normal samples. (PNG 725 kb)

277_2023_5409_Fig9_ESM.png
Fig. S3 The expression level of CCDC50 V1 and V2 in DLBCL (TCGA). The expression level of each exon (A) and junction (B) of CCDC50 in DLBCL patients. (C) The expression level of CCDC50 variants and the proportion of each variant in DLBCL patients. (PNG 219 kb)
277_2023_5409_MOESM8_ESM.csv
Supplementary file5 The raw data of Fig. 1F. (CSV 12 kb)
277_2023_5409_MOESM9_ESM.xlsx
Supplementary file6 The raw data of Fig. 1I. (XLSX 10 kb)
277_2023_5409_MOESM10_ESM.xlsx
Supplementary file7 The raw data of Fig. 2C.(XLSX 1029 kb)
277_2023_5409_MOESM11_ESM.xlsx
Supplementary file8 The raw data of Fig. S2C. (XLSX 11 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gong, Y., Tong, H., Yu, F. et al. CCDC50, an essential driver involved in tumorigenesis, is a potential severity marker of diffuse large B cell lymphoma. Ann Hematol 102, 3153–3165 (2023). https://doi.org/10.1007/s00277-023-05409-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-023-05409-w