Abstract
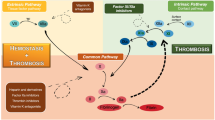
COVID-19 is characterized by a predominantly prothrombotic state, which underlies severe disease and poor outcomes. Imbalances of the gut microbiome have been linked with abnormal hemostatic processes. Understanding the relationship between the gut microbiome and abnormal coagulation parameters in COVID-19 could provide a novel framework for the diagnosis and management of COVID-related coagulopathies (CRC). This cross-sectional study used shotgun metagenomic sequencing to examine the gut microbiota of patients with CRC (n = 66) and compared it to COVID control (CCs) (n = 27) and non-COVID control (NCs) (n = 22) groups. Three, 1, and 3 taxa were found enriched in CRCs, CCs, and NCs. Next, random forest models using 7 microbial biomarkers and differential clinical characteristics were constructed and achieved strong diagnostic potential in distinguishing CRC. Specifically, the most promising biomarker species for CRC were Streptococcus thermophilus, Enterococcus faecium, and Citrobacter portucalensis. Conversely, Enterobacteriaceae family and Fusicatenibacter genus are potentially protective against CRC in COVID patients. We further identified 4 species contributing to 20 MetaCyc pathways that were differentially abundant among groups, with S. thermophilus as the main coding species in CRCs. Our findings suggest that the alterations of gut microbiota compositional and functional profiles may influence the pathogenesis of CRC and that microbiota-based diagnosis and treatment could potentially benefit COVID patients in preventing and alleviating thrombosis-related clinical outcomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coagulopathy is among the most prominent features of COVID-19 with a predominance of a prothrombotic state. The laboratory signature of COVID-19-associated coagulopathy (CRC) includes elevated fibrinogen (FIB) levels, marked increases in D-dimer concentration, relatively modest thrombocytopenia, and prolongation of the prothrombin time (PT) or/and activated thromboplastin time (APTT) [1, 2]. These distinct hematological features are found associated with disease severity and poor outcomes, and prophylactic and therapeutic anticoagulation therapies have been reported to improve the prognosis of COVID-19 patients [3].
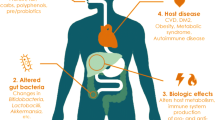
The gut microbiome plays important roles in host immune homeostasis, metabolism, infection, and hemostatic processes [4,5,6,7]. Serum gut microbiome-derived metabolites, namely trimethylamine N-oxide (TMAO) and phenylacetylglutamine (PAGln) have been established to be predictive of thrombotic cardiovascular diseases [8, 9]. Gut commensals could also influence platelet function [10], VWF synthesis [11], and Vitamin K2 metabolism [12]. When the gut epithelial barrier is impaired by stress such as inflammation and viral infection, gut microbes, microbial products (e.g., lipopolysaccharides [LPS]), and metabolites (e.g., TMAO) translocate into the portal and then systemic circulation with resultant bacteremia and endotoxemia and potentially disseminated intravascular coagulation (DIC) [13]. Gastrointestinal involvement is evident in COVID-19 patients [14]. Furthermore, significant alterations in gut microbiota composition and functionality have been reported in previous studies, as well as their association with disease severity, dysfunctional immune responses, and impaired capacity for short-chain fatty acid (SCFA) and branched chain amino acid (BCAA) synthesis [15,16,17]. Therefore, it is possible that in SARS-CoV-2 infection, gut barrier and gut microbiome disruption work synergistically with striking inflammatory response to promote the prothrombotic state [13, 18].
However, to the best of our knowledge, no study has investigated the gut microbiome compositional and functional profiles related to CRC. Therefore, this work aims to map the gut microbiome profiles of COVID-19 patients with abnormal coagulation parameters, examine their relationships, and evaluate the potential clinical discriminatory power of the discovered bacterial biomarkers in predicting CRC.
Materials and methods
Study subject and fecal sample collection
This study was approved by the ethics committee of the Wuhan Union Hospital (NO. 0033), and all participants signed informed consent forms.
This study involved 93 patients with COVID-19 hospitalized with laboratory-confirmed SARS-CoV-2 infection and 22 non-COVID individuals (non-COVID controls, NCs). SARS-CoV-2 infection was diagnosed by reverse-transcriptase polymerase chain reaction (RT-PCR) assay using respiratory tract samples. The participating COVID-19 patients were admitted to the Wuhan Union Hospital from January to March 2020. Additionally, NCs were otherwise healthy patients exhibiting respiratory symptoms who were confirmed with no COVID infection recruited from the hospital quarantine site in the same time period. COVID-19 disease severity was determined using the diagnostic criteria of the seventh edition of the Diagnostic and Treatment Protocol for COVID-19 in China [19]. Participants’ clinical characteristics, including demographic characteristics (age, gender), comorbidities (hypertension, diabetes, heart diseases, kidney diseases), GI symptoms (diarrhea, vomiting, and anorexia), and antibiotics/probiotics use and nutrition support during hospital stay were collected from the medical records.
Fecal samples were collected during hospital stay (median 24, range 8–20 days after admission) and immediately sent to the laboratory for inactivation at 95°C for 30 min and then sent to the Wuhan Huada Gene Laboratory to test for metagenomics.
Analysis of coagulation profile
Blood samples were collected serially until patient discharge for evaluation of coagulation profiles using automated equipment and standard methods. All laboratory tests, specifically, coagulation tests including PT, aPTT, FIB, and D-dimer, were done in the core laboratory of the Wuhan Union Hospital with standard procedures.
Microbial profiling of fecal samples with metagenomic sequencing
The microbial community DNA was extracted using TIANamp Stool DNA Kit (DP328, Tiangen Biotech) following the manufacturer’s instructions. The integrity of the DNA samples was analyzed via 2% (w/v) agarose gel electrophoresis. Subsequently, DNA libraries were constructed through the processes of end repairing, adding A to tails, purification, and PCR amplification using an Ion Torrent Proton Sequencer (Life Technologies, Carlsbad, California). Quality control of the DNA libraries was carried out using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California) to assess the DNA concentration and fragment size. Qualified DNA libraries were sequenced on the BGISEQ platform (MGI BGISEQ-50, BGI Wuhan Clinical Laboratories, BGI-Shenzhen, Wuhan 430074, China).
Raw sequence reads were filtered and quality-trimmed using Trimmomatic v0.39 as follows: (1) trimming low-quality base (quality score < 20), (2) removing reads shorter than 35 base pairs, and (3) removing sequencing adapters. Contaminating human reads were filtered using Kneaddata (reference database: GRCh38/hg38) with default parameters. The microbial taxonomic and functional profiles were extracted using MetaPhlAn3 and HUMAnN3, respectively.
Bioinformatic analysis
Relative abundance data from MetaPhlAn3 were imported into R for downstream analysis. Alpha diversity was assessed using the Shannon diversity index. Beta diversity was estimated by the Bray-Curtis distance and was visualized by principal coordinate (PCoA) and non-metric multidimensional scaling (NMDS) analyses. Adonis test was used to assess overall microbiome structure differences between defined groups. Differential bacterial taxa across groups (COVID-19 patients with abnormal coagulation parameters [CRCs], COVID-19 controls [CCs], NCs) were determined using linear discriminant analysis (LDA) effect size (LefSe) analysis based on an LDA score > 3.0. We used the multivariate association with linear models (MaAsLin) to further identify the associations between microbial features with abnormal coagulation profiles, controlling for clinical confounders. Biomarkers were finally identified as the overlapping taxa among LefSe and MaAsLin analysis results.
Based on differential taxa and clinical variables, random forest classifiers were trained on data from the discovery cohort. Five-fold cross-validation was used to evaluate the performance of the predictive model. In the cross-validation error curve, the number of variables at the lowest cross-validation error was used to construct the prediction model. We evaluated the discriminating efficacy of the sets of biomarkers in the discovery and validation cohort using the receiver operating characteristic (ROC) curve and calculated the multiclass area under the curve (AUC) to estimate the overall diagnostic value [20] and separate AUCs. In general, an AUC of 0.6 to 0.70 indicates that the predictive ability of the model is poor, 0.7 to 0.8 is considered acceptable, and 0.8 to 0.9 is considered excellent [21].
Statistical analysis
Continuous variables were described as mean values with standard deviation (SD), and categorical variables were described using frequencies with percentages. Student t-tests and chi-square tests were used to detect differences in clinical characteristics among groups. The Kruskal-Wallis test and Wilcoxon rank-sum test were used for multiple-group and pairwise comparisons. The rank biserial correlation and point biserial correlation were used to assess the relationship between the presence of abnormal coagulation parameters, disease severity, and biomarker abundances.
False discovery rate (FDR, Benjamini–Hochberg) was applied to correct for multiple comparisons. A two-sided p < 0.05 was considered significant. All calculations were performed using R v4.1.2 and SPSS 22.0.
Results
Demographic and clinical characteristics of the participants
Among the 115 participants, 63 received empirical antibiotics or probiotics during hospitalization. Sixty-six patients presented with one or more abnormal coagulation functions during hospitalization, among which 49 (74.2%) presented with D-dimer elevation, 42 (63.6%) with FIB elevation, 26 (39.4%) with coagulopathy (prolonged PT and/or aPTT), and 13 (19.7%) and 14 (21.2%) with thrombocytopenia and thrombocytosis. The mean ages of CRCs, CCs, and NCs were 62.09 ± 10.78, 54.89 ± 12.04, and 45.41 ± 10.32 years, respectively; 58.5% (n = 38) and 32.0% (n = 8) of CRCs and CCs, respectively, had underlying comorbidities (p = 0.024). All participants presented with respiratory symptoms, but 43 of them also had GI symptoms, including 9 that had diarrhea and 5 that had vomiting at presentation. Significantly more participants in the CRC group presented with GI symptoms at admission (p = 0.04). None of the patients developed GI symptoms when their stool samples were collected. Additionally, 11 patients (oral nutrition support, n = 8; enteral tube feeding, n = 3), all in the CRC group, received nutrition support during hospitalization (Table 1).
Using rank-biserial correlation tests, we found that abnormal coagulation parameters (D-dimer elevation, prolonged PT and/or aPTT, FIB elevation, and thrombocytopenia) were mild-to-moderately associated with COVID-19 disease severity (r = 0.24–0.48, p < 0.05) (Supplementary Fig S1).
Bacterial diversity increased in the guts of COVID-19 patients compared with non-COVID controls
We first examined the Shannon index across CRCs, CCs, and NCs. Curiously, the taxa diversity at the species level in CRCs was similar to CCs (p = 0.18), but the diversity of COVID patients was significantly higher than that in NCs (p < 0.01) (Fig. 1a). To determine whether the overall gut microbial composition of CRCs, CCs, and NCs was different, we examined different β-diversity measures. In the PCoA plots, these groups were clustered separately, indicating that their compositions were significantly different; similar results were observed in NMDS plots (Fig. 1b–c). These conclusions were also confirmed by permutational multivariate analysis of variance (p = 0.016). Subgroup analyses in cohorts with and without comorbidities, naïve to and treated with antibiotics and probiotics, with and without GI symptoms at admission, and different types of nutrition support, similar diversity, and composition differences were found, except for CRCs receiving nasogastric enteral feeding as nutrition support, which harbored gut bacteria with much lower diversity compared with CCs (p = 0.026) (Supplementary Fig 2-5).
Comparisons of gut bacterial diversity and composition in CRCs, CCs, and NCs. Microbial α-diversity illustrated using a Shannon index plot and β-diversity illustrated using b NMDS plot and c PCoA plot. The gut bacterial composition of CRCs, CCs, and HCs on d phylum level and e genus level. CRC, COVID-related coagulopathy group; CC, COVID controls; NC, non-COVID controls
Altered microbial composition in the guts of COVID-19 patients with abnormal coagulation parameters
The gut microbiome of CRCs was dominated by Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Verrucomicrobia, and Fusobacteria at the phylum level. The abundance of Fusobacteria phylum was significantly enriched in the CRC group (mean 0.55%), which was much higher than that in CCs (mean 0.07%), whereas in the NC group, Fusobacteria was not detected. Also, a marked decrease in the Firmicutes/Bacteroidetes ratio could be observed in COVID patients (Fig. 1d). Actinobacteria in COVID patients also increased. On the genus level, Bacteroides, Blautia, Bifidobacterium, Ruminococcus, and Eubacterium were enriched in COVID patients, while Veillonella, Akkermansia, and Erysipelatoclostridium were enriched in NCs. It is noteworthy that Streptococcus overexpression was specific to the CRC group (mean 11.41%) compared with CCs (mean 3.53) and NCs (2.87%). Also, an underrepresentation of Escherichia and Enterococcus could be found in CCs (Fig. 1e).
We then applied one-against-all comparisons using LEfSe analyses between CRCs, CCs, and NCs. From phylum to species level, LEfSe analysis identified 27, 7, and 8 biomarker taxa (LDA scores > 3, adjusted p-values [p-adj] < 0.05) in CRCs, CCs, and NCs, respectively (Supplementary Table S1, Supplementary Figure S6). The use of antibiotics and probiotics and different types of nutrition support were known factors that could influence the gut microbiota [22,23,24,25]. To de-confound the potential effects of clinical variables, factors significantly different between CRCs and CCs (i.e., comorbidities and GI symptoms at admission), as well as antibiotic/probiotic treatment and nutrition support statuses, were included in the MaAsLin analysis, which further identified 19 significant associations (p-adj < 0.05) between the relative abundances of 18 taxa and coagulation profiles (Supplementary Table S2).
Finally, the two analyses yielded 3, 1, and 3 overlapping biomarkers, for CRC, CC, and NC, separately. Specifically, the enrichment of Streptococcus thermophilus (LDA = 4.53, p-adj = 0.004), Enterococcus faecium (LDA = 4.37, p-adj = 0.005), and Citrobacter portucalensis (LDA = 3.07, p-adj = 0.035) showed prominent abilities to discriminate CRCs from CCs and NCs. Fusicatenibacter genus (LDA = 3.74, p-adj = 0.038) turned out to be the only biomarker for the CC group. Enterobacteriaceae (LDA = 4.56, p-adj = 0.0499) was increased in relative abundance from class to family level in the NC group, compared with CRC and CC groups (Fig. 2a).
Biomarkers for CRCs, CCs, and NCs, identified by overlapping LEfSe and MaAsLin results. Biomarkers for CRCs were identified by overlapping LEfSe.CRC with MaAsLin.NC.neg and/or MaAsLin.CRC.pos; biomarkers for CCs were identified by overlapping LEfSe.CC with MaAsLin.NC.neg and/or MaAsLin.CRC.neg; and biomarkers for NCs were identified by overlapping LEfSe.NC with MaAsLin.NC.pos and/or MaAsLin.CRC.neg. LDA, linear discriminant analysis; LefSe, LDA effect size analysis; MaAsLin, multivariate association with linear model; pos, positive B-coefficients; neg, negative B-coefficients; CRC, COVID-related coagulopathy group; CC, COVID controls; NC, non-COVID controls
Additionally, partial correlation analyses controlling for clinical characteristics (i.e., comorbidities, GI symptoms at admission, antibiotic/probiotic treatment, and nutrition support statuses) were performed to show the associations between the above biomarker taxa and specific coagulation profiles. Enterobacteriaceae, Enterobacterales, and Gammaproteobacteria showed moderate to strong positive correlations (r = 0.59–0.71, p < 0.05) with normal D-dimer, PT and/or aPTT, and FIB parameters, while S. thermophilus and E. faecium were associated with platelet abnormalities (r = 0.08–0.27, p < 0.05) (Fig 3).
Identification of a classifier for COVID-19 patients with abnormal coagulation functions based on the microbial candidates and clinical characteristics
In the discovery cohort, a random forest classifier model including 92 samples was constructed to evaluate the potential of gut microbial and clinical markers as a prediction tool for CRC. Through 5-fold cross-validation of the random forest model, 7 taxa and 4 clinical variables were selected as the optimal marker set (Fig. 4a). Mean decrease accuracy was used to calculate the variable importance of the variables (Fig. 4b). In the discovery cohort, the multiclass AUC value was 100%. In the validation cohort, the multiclass AUC reached 93.5%. Specifically, the AUC value was 82.7% (95% CI: 59.1–100.0%) between CRCs and CCs, 100% (95% CI: 100–100.0%) between CRCs and NCs, and 100% (95% CI: 100–100%) between CCs and NCs (Fig 4c), indicating that the classifier model based on microbial markers reached excellent diagnostic potential in discriminating between CRCs, CCs, and NCs. The classifier combining microbial and clinical information significantly outperformed classifiers based on only biomarker taxa or differential clinical characteristics (Supplementary Figures S7-S8).
Diagnostic potential of microbial and clinical biomarkers for CRC. a Cross-validation curve identified a total of 7 taxa and 4 clinical variables by the RF model; b variable importance measured by mean decrease in accuracy; c ROC curve and AUC calculated based on a test set of the validation cohort (red line, CRCs vs. CCs; blue, CRCs vs. NCs; green, CCs vs. NCs). RF, random forest; ROC, receiver operating characteristic; AUC, area under the curve; CRC, COVID-related coagulopathy group; CC: COVID controls; NC, non-COVID controls; GI, gastrointestinal
Crucial pathways related to COVID-19-related coagulopathies
We retrieved 546 MetaCyc pathways from the functional profiles of each sample to analyze crucial microbial pathways related to CRC. Among the groups, 18 pathways were significantly different (Kruskal-Wallis test, p < 0.05). These pathways were involved in amino acid, carbohydrate, lipid, nucleoside and nucleotide metabolism, and enzyme biosynthesis. Specifically, L-arginine and L-methionine biosynthesis pathways were prominently strengthened in the gut microbiota in COVID patients (Fig. 5a). Four species, namely S. thermophilus, E. faecium, Escherichia coli, and Ruminococcus gnavus, contributed within 7 pathways (Kruskal-Wallis test, p < 0.05). S. thermophilus showed significant functional shifts after COVID infection, including L-methionine biosynthesis and purine nucleotide biosynthesis and degradation (Wilcoxon rank-sum test, p < 0.001) (Fig. 5b).
Discussion
As an important component of human immunity and metabolism, gut microbes have attracted extensive attention in COVID-19. These studies have found that there are significant immediate, as well as prolonged changes in the gut microbiota of COVID-19 patients: decreased diversity, loss of commensal bacterial taxa and functions known to have immunomodulatory activity, such as short-chain fatty acid (SCFA) production, and increased abundance of opportunistic pathogens related with inflammatory reactions [26, 27]. This study is the first attempt to investigate the role of the gut microbiome in COVID-19-related coagulopathy. Additionally, we employed metagenomic shotgun sequencing for comprehensive analyses, which has more power to identify less abundant taxa than 16S sequencing [28]. In our study, we identified differential CRC microbial markers and constructed a reliable random forest model combining microbial biomarkers and clinical variables using 5-fold cross-validation, which showed strong diagnostic potential in distinguishing CRCs from CCs and NCs. Based on the changes in microbial species, some important metabolic pathways were upregulated or downregulated in COVID-19 patients and those with CRC.
Dysbiosis, disturbance or imbalance in the gut microbiota, is generally accompanied by reduced diversity and an increase in pathogenic microbes. However, in our study, COVID patients (CRCs and CCs alike), harbored a more diverse microbiota than NCs, which was contradictory to previous studies, which often pointed to non-significant differences or decreased diversity in COVID patients [15, 26, 29]. This could potentially be explained by using otherwise healthy individuals exhibiting respiratory symptoms who were confirmed with no COVID infection from the hospital quarantine site as NCs. Decreased Firmicutes/Bacteroidetes ratio, a widely accepted indicator of gut inflammation [30], was observed in COVID patients, supporting previous studies [31]. Additionally, a decrease in the Firmicutes/Bacteroidetes ratio similar to some systemic inflammatory conditions has been observed in primary immune thrombocytopenia (ITP) patients, indicating the presence of the association between platelet activation and intestinal microbiota [32].
Consistent with existing clinical evidence, CRC correlates with COVID-19 disease severity [33], which warrants deeper insights into its pathogenesis and targeted prevention and treatments. Using LEfSe and MaAsLin, our study pinpointed several opportunistic pathogens as biomarkers for CRC.
Curiously, S. thermophilus, a homofermentative regarded as a symbiont harboring anti-inflammatory properties [34], showed striking overrepresentation in CRC fecal samples, even after controlling for probiotic use and other clinical factors. Furthermore, it correlated with platelet abnormalities. Its changed functional profile could be one of the explanations for its prothrombotic properties. Focusing on amino acid-related pathways, we found that L-methionine biosynthesis via O-phospho-homoserine and methionine to cysteine transsulfuration increased in a gradient manner from NCs, CCs, to CRCs. A metagenome-wide association study found that the abundance of L-methionine synthesis was significantly higher in individuals with carotid atherosclerotic plaques [35]. We hypothesize the central role of sulfur-containing amino acid methionine in CRC could be related to increasing homocysteine, as L-methionine can be converted to homocysteine directly or through S-adenosyl-L-methionine, as shown by the elevated abundance of the above pathways via the intermediate metabolite L-homocysteine. Homocysteine-related thrombotic diseases have been intensively investigated over the past decades. Hyperhomocysteinemia has been found to cause platelet apoptosis and subsequently enhance thrombogenicity; it also causes a decrease of the bioavailability of endothelial nitric oxide (NO), which inhibits of platelet aggregation [36]. Clinical evidence has introduced high levels of circulating homocysteine as a strong risk factor for thromboembolism, influencing platelet reactivity [37, 38]. Indeed, homocysteine has been identified as a biomarker for COVID progression and severity [39], while another article contested that monitoring homocysteine levels is necessary for preventing pulmonary embolism in COVID patients [40]. Our study added to the literature a microbiota perspective.
E. faecium is a facultatively anaerobic opportunistic pathogen with extensive antibiotic resistance, which could cause severe nosocomial infections, especially in critically ill patients in intensive care units (ICU). It is an invasive species that could pass through the intact mucosal epithelium and enter the circulation; when the gut barrier is compromised, its translocation could lead to sepsis and a subsequent prothrombotic state [41]. Our study found a weak correlation between E. faecium and abnormal platelet counts. Recently, PrpA, a thermosensitive surface protein in E. faecium from clinical samples, was found to be able to bind to fibrinogen, fibronectin, and platelets, especially activated platelets [42], indicating a potential role of E. faecium in abnormal clotting.
We proposed that probiotic strains of Enterobacteriaceae family could potentially be utilized to resist infection [43] and protect patients against CRC, especially D-dimer and fibrin elevation. Another beneficial organism was Fusicatenibacter genus, which was generally depleted in patients with COVID in different stages (acute, convalescence, and post-convalescence) [44]. A preliminary study showed that Fusicatenibacter saccharivorans exhibited anti-inflammatory properties in ulcerative colitis by inducing the production of anti-inflammatory cytokines, enhancement of intestinal epithelial barrier functions, and producing SCFA [45]. Similar mechanisms could be implicated in the protective role of the enrichment of this flora in patients with COVID who did not develop CRC.
Using the random forest classifier, we demonstrated the prospects of targeted biomarkers of the gut microbiome combined with clinical information serving as a reliable diagnostic tool for CRC. This new diagnostic tool can be used as a supplement to direct prophylactic and therapeutic anticoagulation therapies in COVID-19 patients, especially those with a high risk of developing critical conditions or with severe comorbidities.
Although this study provides valuable insights into CRC from a microbiome perspective, it has several limitations that should be addressed in future studies. Firstly, it is a single-center study involving a small number of samples. Secondly, this study would be greatly enhanced by metabolomics and proteomics analyses. We concede that large prospective matched cohort studies are needed to verify the diagnostic efficacy, and more physiological parameters and regular collection of CRC patients’ feces samples should be added to future study designs.
Data availability
The data that support the findings of this study are available upon request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
Levi M, Thachil J, Iba T, Levy JH (2020) Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol 7:e438–ee40
McBane RD 2nd, Torres Roldan VD, Niven AS, Pruthi RK, Franco PM, Linderbaum JA et al (2020) Anticoagulation in COVID-19: A systematic review, meta-analysis, and rapid guidance from Mayo Clinic. Mayo Clin Proc 95:2467–2486
Zhang S, Li Y, Liu G, Su B (2021) Intermediate-to-therapeutic versus prophylactic anticoagulation for coagulopathy in hospitalized COVID-19 patients: a systemic review and meta-analysis. Thromb J 19:91
Fan Y, Pedersen O (2021) Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19:55–71
Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z et al (2016) Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165:111–124
Lin L, Zhang J (2017) Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol 18:2
Jansen VL, Gerdes VE, Middeldorp S, van Mens TE (2021) Gut microbiota and their metabolites in cardiovascular disease. Best Pract Res Clin Endocrinol Metab 35:101492
Li XS, Obeid S, Klingenberg R, Gencer B, Mach F, Räber L et al (2017) Gut microbiota-dependent trimethylamine N-oxide in acute coronary syndromes: a prognostic marker for incident cardiovascular events beyond traditional risk factors. Eur Heart J 38:814–824
Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM et al (2020) A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell 180:862–77.e22
Zhang X, Zhang X, Tong F, Cai Y, Zhang Y, Song H et al (2022) Gut microbiota induces high platelet response in patients with ST segment elevation myocardial infarction after ticagrelor treatment. eLife 11:e70240
Jäckel S, Kiouptsi K, Lillich M, Hendrikx T, Khandagale A, Kollar B et al (2017) Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via Toll-like receptor-2. Blood 130:542–553
Daisley BA, Koenig D, Engelbrecht K, Doney L, Hards K, Al KF et al (2021) Emerging connections between gut microbiome bioenergetics and chronic metabolic diseases. Cell Reports 37:110087
Hasan RA, Koh AY, Zia A (2020) The gut microbiome and thromboembolism. Thromb Res 189:77–87
Synowiec A, Szczepański A, Barreto-Duran E, Lie LK, Pyrc K (2021) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a systemic infection. Clin Microbiol Rev 34
Zhang F, Wan Y, Zuo T, Yeoh YK, Liu Q, Zhang L et al (2021) Prolonged impairment of short-chain fatty acid and Lisoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology
Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY et al (2020) Alterations in fecal fungal microbiome of patients with covid-19 during time of hospitalization until discharge. Gastroenterology 159:1302–10.e5
Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H et al (2020) Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 159:944–55.e8
Gu SX, Tyagi T, Jain K, Gu VW, Lee SH, Hwa JM et al (2021) Thrombocytopathy and endotheliopathy: Crucial contributors to COVID-19 thromboinflammation. Nat Rev Cardiol 18:194–209
National Health Commission of the People’s Republic of China, National Administration of Traditional Chinese Medicine. Guideline for Diagnosis and Treatment of COVID-19 (7th version). 2020.
Mandrekar JN (2010) Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 5:1315–1316
Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy JM, Dequenne I et al (2019) How probiotics affect the microbiota. Front Cell Infect Microbiol 9:454
Becattini S, Taur Y, Pamer EG (2016) Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22:458–478
Whelan K, Judd PA, Tuohy KM, Gibson GR, Preedy VR, Taylor MA (2009) Fecal microbiota in patients receiving enteral feeding are highly variable and may be altered in those who develop diarrhea. Am J Clin Nutr 89:240–247
Katagiri S, Shiba T, Tohara H, Yamaguchi K, Hara K, Nakagawa K et al (2019) Re-initiation of oral food intake following enteral nutrition alters oral and gut microbiota communities. Front Cell Infect Microbiol 9:434
Sarkar A, Harty S, Moeller AH, Klein SL, Erdman SE, Friston KJ et al (2021) The gut microbiome as a biomarker of differential susceptibility to SARS-CoV-2. Trends Mol Med 27:1115–1134
Hand DJ, Till RJ (2001) A simple generalisation of the area under the roc curve for multiple class classification problems. Mach Learn 45:171–186
Zuo T, Wu X, Wen W, Lan P (2021) Gut microbiome alterations in COVID-19. Genom Proteom Bioinform 19:679–688
Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A (2021) Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep 11:3030
Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY et al (2021) Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70:698–706
Stojanov S, Berlec A, Štrukelj B (2020) The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms:8
Moreira-Rosário A, Marques C, Pinheiro H, Araújo JR, Ribeiro P, Rocha R et al (2021) Gut microbiota diversity and c-reactive protein are predictors of disease severity in COVID-19 patients. Front Microbiol 12:705020
Liu C, Cheng L, Ji L, Li F, Zhan Y, Wu B et al (2020) Intestinal microbiota dysbiosis play a role in pathogenesis of patients with primary immune thrombocytopenia. Thromb Res 190:11–19
Arachchillage DRJ, Laffan M (2020) Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18:1233–1234
Martinović A, Cocuzzi R, Arioli S, Mora D (2020) streptococcus thermophilus: to survive, or not to survive the gastrointestinal tract, that is the question! Nutrients 12
Kurilshikov A, ICLvd M, Chen L, Bonder MJ, Schraa K, JHW R et al (2019) Gut microbial associations to plasma metabolites linked to cardiovascular phenotypes and risk. Circulation Research 124:1808–1820
Dionisio N, Jardín I, Salido GM, Rosado JA (2010) Homocysteine, intracellular signaling and thrombotic disorders. Curr Med Chem 17:3109–3119
Lupi-Herrera E, Soto-López ME, Lugo-Dimas AJ, Núñez-Martínez ME, Gamboa R, Huesca-Gómez C et al (2019) Polymorphisms C677T and A1298C of MTHFR gene: homocysteine levels and prothrombotic biomarkers in coronary and pulmonary thromboembolic disease. Clin Appl Thromb Hemost 25:1076029618780344
Verdoia M, Rolla R, Negro F, Tonon F, Pergolini P, Nardin M et al (2020) Homocysteine levels and platelet reactivity in coronary artery disease patients treated with ticagrelor. Nutr Metab Cardiovasc Dis 30:292–299
Ponti G, Maccaferri M, Ruini C, Tomasi A, Ozben T (2020) Biomarkers associated with COVID-19 disease progression. Crit Rev Clin Lab Sci 57:389–399
Ibrahimagić O, Smajlović D, Dostović Z, Kunić S, Šehanović A, Kojić B (2020) Comment on an article: "Coagulopathy in COVID-19". J Thromb Haemost 18:3381–3382
Krawczyk B, Wityk P, Gałęcka M, Michalik M (2021) The many faces of enterococcus spp.-commensal, probiotic and opportunistic pathogen. Microorganisms:9
Guzmán Prieto AM, Urbanus RT, Zhang X, Bierschenk D, Koekman CA, van Luit-Asbroek M et al (2015) The N-terminal domain of the thermo-regulated surface protein PrpA of Enterococcus faecium binds to fibrinogen, fibronectin and platelets. Sci Rep 5:18255
Velazquez EM, Nguyen H, Heasley KT, Saechao CH, Gil LM, Rogers AWL et al (2019) Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nature Microbiology 4:1057–1064
Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J et al (2022) Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 71:222–225
Takeshita K, Mizuno S, Mikami Y, Sujino T, Saigusa K, Matsuoka K et al (2016) A single species of clostridium subcluster xiva decreased in ulcerative colitis patients. Inflamm Bowel Dis 22:2802–2810
Funding
This study was supported by the Department of Science and Technology of Hubei Province, grant numbers 2020FCA014, 2020BHB017, XGFY2020000027, and YFYB2020001694. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.S., T.B., and X.H.; data curation, T.B.; formal analysis, Y.C.; funding acquisition, T.B.; investigation, X.L, C.Y., E.W., C.L., Y.J., L.Z., Y.M., Y.J., L.Y., and T.B.; methodology, Y.C. and T.B.; project administration, T.B.; resources, X.L, C.Y., E.W., C.L., Y.J., L.Z., Y.M., Y.J., L.Y., B.S., J.Q., X.Z., L.R., H.E., and T.B.; software, Y.C., X.Z., and L.R.; supervision, T.B.; validation, T.B., B.S., J.Q., X.Z., H.E., and L.R.; visualization, Y.C.; writing—original draft, Y.C.; writing—review and editing, B.S., J.Q., J.S., T.B., H.E., and X.H. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008. Informed consent was obtained from all patients for being included in the study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Y., Li, X., Yu, C. et al. Gut microbiome alterations in patients with COVID-19-related coagulopathy. Ann Hematol 102, 1589–1598 (2023). https://doi.org/10.1007/s00277-023-05186-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-023-05186-6