Abstract
Species can either maintain a certain social organization in different habitats or show different social organizations in similar habitats. The reasons underlying this variability are not always clear but might have consequences for population dynamics, especially under changing environmental conditions. Among mammals, the primate genus Microcebus lives in small groups of closely related females, derived from female philopatry and dispersed males, as illustrated by the well-studied Microcebus murinus. Here, we studied the genetic structure of a population of the congeneric Microcebus griseorufus, inhabiting three adjacent habitats with different resource availabilities. In order to learn more about the plasticity of the species’ social organization under these different conditions, we analyzed the spatial arrangement of mitochondrial haplotypes of 122 individuals. The study revealed high haplotype diversity and a pronounced difference in spatial distribution between the sexes. Females exhibited spatial aggregation of haplotypes, suggesting a system of female philopatry and matrilines, similar to M. murinus. Male haplotypes were dispersed, and males were more likely to carry rare haplotypes, indicating higher dispersal activity. These findings hint towards the unity of the social organization across the genus Microcebus, suggesting a phylogenetic origin of the social organization. Yet, with decreasing resources, the clustering of female haplotypes declined and approached a random distribution in the marginal habitat, with cluster sizes correlating with resource availability as predicted by the socioecological model. Our study supports the notion that social organization is shaped by both phylogenetic origin and ecological conditions, at least in these small primates.
Significance statement
Impacts of habitat degradation are mostly described in terms of changes in population densities in relation to the reduction of resources. This neglects the possible effects of altered social organizations due to declining resources or population densities. Using a genetic sampling of three subpopulations of mouse lemurs in Madagascar along a gradient of food availability up to the limit of the species’ ecological tolerance, we show that their social organization consisting of spatial clusters of closely related females and overdispersed males converges towards random spatial distributions of both sexes with declining food availability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vast majority of nonhuman primate populations are declining due to the degradation and destruction of their habitats, hunting, and climate change (Estrada et al. 2017). Studies on habitat degradation focus on identifying resources that are important for the persistence of species in degraded habitats. In most cases, “importance” is defined through correlations of specific resource densities with population densities of primates as well as of other taxa (e.g., Johns and Skorupa 1987; Chapman et al. 2000; Irwin et al. 2010; Peres et al. 2010; Sodhi et al. 2010; Steffens et al. 2023). Conclusions about the viability of populations based on the focus on resources neglect behavioral responses and consequences of habitat change on social systems and life histories that could either provide solutions or reflect constraints, leading to intra-specific variation in social organization due to social flexibility (Lott 1984; Schradin 2013; Schradin et al. 2018; Ozgul et al. 2023). The socioecological model for the evolution of different social systems, made popular by Krebs and Davies (1978) and refined continuously thereafter, has provided a useful framework for the study of the social and mating systems of animals. The basic assumption is that females and males evolved social systems that maximize their reproductive success, sometimes with diverging sex-specific interests (Trivers 1972; Cassini 2021). Due to their impressive diversity of social systems and a large number of case studies, primates have often served as examples to test predictions of the model (Crook 1964; Emlen and Oring 1977; Wrangham 1980; Terborgh and Janson 1986; Dunbar 1988; van Schaik 1989; Goldizen 1990; Sterck et al. 1997; Kappeler and van Schaik 2002; Koenig and Borries 2009; Clutton-Brock and Janson 2012; Koenig et al. 2013). The socioecological model uses ecological parameters such as resource distribution in space and time to explain the social organization such as spatial distributions and group size. Simplified, the spatial distribution and relationships of females are determined by food availability with associated within-group and between-group feeding competition and risk avoidance, while male distribution and relationships are mainly based on the distribution of mating opportunities (van Schaik 1983; Dunbar 1988; Janson 1992). Where resources are not worth defending, females can be expected to disperse. Males are expected to compete among themselves for receptive females and should adjust their mating strategy to optimize access to receptive females. If females are spatially clumped and exhibit non-synchronized estrus cycles, the chances of monopolization of females are high. When females are dispersed, and their estrus cycles are synchronized, the potential to monopolize females is reduced, leading to dispersed males (Ims 1988; Schwagmeyer 1988). While the socioecological model has provided a useful framework for studies on the driving forces of sexual selection, many findings are inconsistent with the model’s prediction, arguing that other factors or phylogenetic constraints might be more important than ecological ones and that optimization has to be considered a concept rather than a state that can be achieved (Pulliam and Caraco 1984; Brockman 2001; Koenig and Borries 2009; Clutton-Brock and Janson 2012; Koenig et al. 2013; Strier 2017; Fischer et al. 2019).
Evolutionary processes and aspects of social organization, such as dispersal and breeding strategies, affect the genetic structure of a population (Hamilton 1964; Wright 1965; Wilson 1975; Ross 2001). Because of this association between social organization and genetic structure, information on the genetic structure is essential in helping us understand social behavior and analyze social systems (Kappeler and van Schaik 2002). Genetic data such as measurements of genetic similarity can provide insights into social systems that are not attainable through observational methods alone (Wimmer et al. 2002), especially in solitary and nocturnal species, whose social structures are difficult to observe.
The different species of mouse lemurs (genus Microcebus spp.) are nocturnal solitary foragers (Kappeler et al. 2022), whose females are hypothesized to exhibit spatial philopatry with the males dispersing. Female philopatry results in spatial clusters of closely related females with highly overlapping home ranges (Radespiel et al. 1998, 2001; Wimmer et al. 2002; Génin 2008, 2010; Schliehe-Diecks et al. 2012; Génin and Rambeloarivony 2018). Yet, the genus consists of 20+ species, some with very small distributional ranges. These small ranges suggest that the different species differ in specific traits, and question whether or not all species can be assigned to the same ecological and behavioral categories. Studies of different aspects of their social organization are restricted to a handful of species and the genetic structure of their social organization has been described in detail only for Microcebus murinus (Radespiel et al. 2001; Wimmer et al. 2002; reviewed by Kappeler et al. 2022). Different species of Microcebus living in sympatry with M. murinus, share some of the characteristics of the social organization of M. murinus but deviate in others (Microcebus berthae: Dammhahn and Kappeler 2005; Microcebus ravelobensis: Weidt et al. 2004). The differences between sympatric species and the small geographic ranges of others raise the question of whether the genetic correlates described for the social organization of M. murinus can be extrapolated to other species of Microcebus and whether or not the organization is altered intra-specifically by different environmental conditions.
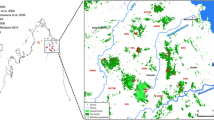
The Grey-brown Mouse Lemur, Microcebus griseorufus (Kollman, 1910), occurs in the spiny forest in the south and southwest of Madagascar (Rasoazanabary 2004; Mittermeier et al. 2010). The vegetation types inhabited by the species range from dry forest resembling a real forest with a closed canopy during the wet season, to the spiny forest and xerophytic bush with few trees and discontinuous vegetation cover (Moat and Smith 2007; Ratovonamana et al., 2011), the latter seemingly representing the dry limit for the species (Bohr et al. 2011; Steffens et al. 2017). In the dry and spiny forest, the species forms population nuclei where related females live in close proximity and defend resources that provide food reliably against other animals (Génin 2008, 2010; Génin and Rambeloarivony 2018). These population nuclei might be equivalent to the spatial clusters of closely related females in M. murinus. According to the socioecological model, the number of females defending these food resources should decline with the declining size of food patches due to increased intra-group competition. Eventually, the patches should become too small to warrant defense. The question is then, whether or not the social organization of clustered females collapses or is maintained, and might have come about in the first place, for other reasons, such as phylogenetic inertia. For the present study, we chose a population of M. griseorufus inhabiting three distinct adjacent vegetation types with different food and tree availability, ranging from “lush” dry forest to xerophytic bush that represents the environmental limit to the distribution of M. griseorufus (Fig. 1). The region is characterized by very high unpredictability of rainfall with severe effects on resource availability (e.g., flowers and fruits) (Dewar and Richard 2007; Ratovonamana et al. 2011; Kasola et al. 2020). The unreliability of flowering and fruiting might make it unprofitable to defend specific parts of an area year-round because it is uncertain whether or not the plants in question will actually provide food. Apart from adding to the understanding of the socioecological model, the study also offers the possibility to understand effects of future habitat changes on the social organization with possible consequences on population dynamics (desiccation and habitat degradation due to climatic or anthropogenic impacts; (Hannah et al. 2008; Tadross et al. 2008; Ozgul et al. 2023).
Dry forest on sandy soil (Habitat 1) next to the soda lake and xerophytic bush on calcareous soil (Habitat 3) on the slope and on the plateau. A dry forest on ferruginous soil (Habitat 2) is in the depressions of the plateau to the east (not shown in the photo). Photograph by Yedidya. R. Ratovonamana
In this study, we investigate the genetic structure of M. griseorufus in three distinct, adjacent habitats with different food availability. Towards this end, we supplemented the existing database by Scheel et al. (2015) by sequencing the mitochondrial D-Loop of another 74 individuals. Following the published approach to infer the social organization of Microcebus murinus from genetic information (Radespiel et al. 2001; Wimmer et al. 2002), we use the spatial distribution of mitochondrial haplotypes as a proxy for the social organization. First, we wanted to know whether the social organization described for M. murinus also applies to M. griseorufus. To test this, we analyzed and compared the published population genetic data on M. murinus at the center of its geographic range (Radespiel et al. 2001; Wimmer et al. 2002) with our new genetic data from M. griseorufus, living at the dry environmental limits of the genus (Bohr et al. 2011; Steffens et al. 2017).
Second, we tested whether the social organization remains invariant across the environmental gradient from the dry forest to the even dryer xerophytic bush of the species’ occurrence or whether it changes in relation to resource abundance.
Material and methods
Study site
This study was conducted in the northwestern part of Tsimanampetsotsa National Park in southwestern Madagascar, ca. 85 km south of Toliara (S 24°01′; E 43°44′). Annual rainfall averages around 400 mm with recurrent droughts without rain for several years. The study area is characterized by two different seasons: eight dry months (April–November) and four wet months (December–March) (Ratovonamana et al. 2011).
The study site can be divided into three distinct habitat types (Fig. 1; Table 1), as a result of the topography and edaphic differences (Mamokatra 1999; Andriatsimietry et al. 2009; Rakotondranary et al. 2010; Ratovonamana et al. 2011). These are with declining suitability for M. griseorufus: (1) dry forest on sandy soil (Habitat 1), a 500 m wide area situated between a soda lake to the west and the 40–100 m cliff of the Mahafaly plateau to the east; (2) dry forest on ferruginous soil (Habitat 2), in depressions of the plateau, filled with red sand, and (3) xerophytic bush (spiny bush) on calcareous soil (Habitat 3), covering the slope from the soda lake to the limestone plateau and extending on the plateau towards the east.
Microcebus griseorufus is omnivorous, feeding on gum, fruits, and insects (Génin 2008). Gum is available year-round, provided mostly by large trees, and consumed mainly during the dry season. Fruits are available and consumed especially during the wet season. Abundance data on insects are not available. Food plants have been identified during tracking studies (Bohr et al. 2011; PG unpubl. data). The general vegetation structure of the three vegetation types was described in the study area by a total of 17 30 × 30 m2 plots, recording all plants (herbs to trees; Ratovonamana et al. 2011). Food plants (all plants > 1 m in height) were counted in each habitat in two 5 × 200 m plots, also used to monitor plant phenology (Bohr et al. 2011; Ratovonamana et al. 2011). The xerophytic bush on calcareous soil differs floristically from the other two habitats on the sand. The three habitats differ in the densities of total food plants [ind./ha] and the density of trees with a diameter at breast height (DBH) ≥ 10 cm [ind./ha]. In the dry forest on sandy soil, the total food plant density is 6032 plants/ha, almost twice as high as that of dry forest on ferruginous soil (3121 plants/ha). 1889 food plants/ha are found in the xerophytic bush on calcareous soil. The largest numbers of trees with DBH ≥ 10 cm were found in Habitat 1 (mean ± standard error: 1029 ± 692) and Habitat 2 (1176 ± 823), while in Habitat 3 only roughly one-third of that was recorded (346 ± 280 trees with DBH ≥ 10 cm). Based on the vegetation structure and the number of food plants, we consider the dry forest on sandy soil (Habitat 1) the most suitable habitat for M. griseorufus, followed by the dry forest on ferruginous soil (Habitat 2) and the xerophytic bush on calcareous soil (Habitat 3).
Microcebus captures
We used capture sites as proxies for the spatial association of individuals. Home ranges measure less than 1 ha with males having larger home ranges than females. Males seem to increase their home ranges during the breeding season, though this observation is anecdotal (Bohr et al. 2011; Génin 2008; PG unpubl. data). Female and male home ranges overlap. Since there is no study on M. griseorufus yet that radio-tracked all individuals within a site simultaneously, we do not know the spatial arrangements of the population members. We use the Euclidian distance between capture sites of individuals to interpret possible grouping patterns as detailed below.
Microcebus griseorufus were captured between April 2007 and March 2009 in 6 ha trapping grids (150 m × 400 m) in each of the three habitats. Traps were spaced regularly at 25-m intervals. The trapping grids were ca. 500 m apart. M. griseorufus lemurs were trapped using 119 Sherman live traps (H. B. Sherman Traps, Tallahassee, FL: 7.5 × 7.5 × 30.5 cm) per grid, equipped with ripe bananas as bait and set at a height of 1–2 m shortly before sunset. Traps were checked before sunrise or in the lactation and weaning season at midnight. Traps were set for four nights per trapping session. Captured mouse lemurs were anesthetized with Ketaminhydrochlorid (Ketamin® 100 mg/ml, Parke-Davis, Berlin) and marked individually either by coded ear clipping or a subcutaneous transponder (Trovan® Passive Transponder System, EURO ID, Identifikationssysteme GmbH and CoKG, Weilerswist, Germany). Small tissue samples were taken from the ear and preserved in 70% ethanol for genetic analyses. Betadine was used to disinfect the mouse lemur skin. None of the recaptured individuals showed signs of infection as a result of the treatment. After examination, animals were kept in their Sherman traps to allow recovery from anesthesia. During that time, they were provided with bananas and water. Animals were released at dusk or at the dawn of the trapping day at their capture sites (Scheel et al. 2015). It was not possible to record data blind because our study involved focal animals in the field. Yet, samples for genetic analyses were only numbered and analyzed by people not involved in the fieldwork (fieldwork by PG, YRR, and SJR; genetic analyses by CA, BMS, and TLL). The two datasets were combined after the completion of the genetic analyses.
Age is difficult to estimate accurately in mouse lemurs since young and older adults cannot be reliably distinguished by their outer appearance outside the mating season (Radespiel et al. 2019). Offspring are born between January and February and are sexually mature after the dry season (ca. July–September). Males in the congeneric M. murinus disperse when they are a few months old (Schliehe-Diecks et al. 2012). Under the assumption that life histories are similar in M. griseorufus, all animals caught after August should have been sexually mature. The body mass of the lightest individual caught after August was 27.5 g; thus, we assume that individuals with a body mass > 27 g were adults. Under this assumption, 13 of the 122 individuals had a body mass between 22 and 27 g and thus were potentially juveniles. Yet, the majority of them were females caught in February and April which are more likely to represent adult females after having given birth or being lactating than being juveniles as they could not have achieved a body mass of > 22 g between birth and the capture date. If they would have been juveniles, their inclusion would make the pattern of diverging haplotype distribution between males and females weaker and thus would represent a conservative error.
Molecular methods and data processing
Genomic DNA was extracted prior to this study from the collected tissue samples with a DNeasy Blood and Tissue Kit (Qiagen) following the standard protocol. In addition to the samples already extracted by Scheel et al. (2015), DNA extraction was performed on 74 samples (NCBI accession numbers: OQ605019 - OQ605092). Polymerase chain reaction (PCR) was carried out according to Scheel et al. (2015) using a standard PCR protocol in a 25 μl reaction mixture containing 17.4 μl of nuclease-free water, 2.5 μl of 10× DreamTaq Green Buffer (Thermo Fisher Scientific, Waltham, MA), 1 μl of dNTP mix (5 mM each; Thermo Fisher Scientific, Waltham, MA), 1 μl of each primer (10 μM), 0.1 μl of DreamTaq Green DNA Polymerase (5U/μl; Thermo Fisher Scientific, Waltham, MA) and 2 μl of template DNA.
A fragment of the mitochondrial D-loop containing the hypervariable Region 1 (HV1) was amplified using the primers TsimMgCytbfw2 (forward; 5′-TCGGACAAGTGGC-CTCTAT-3′) and mih1coau (reverse; 5′-GTTATAGTTTCAGGTTAGTCA-3′) (Hapke et al. 2011). DNA amplification of the D-Loop was conducted in a thermocycler (Analytik Jena, Jena, Germany) with the following settings: Initialization at 92.0 °C for 2 min was followed by 35 cycles of denaturation at 92 °C for 40 s, annealing at 55 °C for 60 s and extension at 72 °C for 60 s, with one final elongation step at 72 °C for 5 min.
After PCR, 3 μl of the PCR products was verified on a 1.5% agarose gel and then purified by adding 0.5 μl Exonuclease I (Thermo Fisher Scientific, Waltham, MA) and 1 μl FastAP (Thermo Fisher Scientific, Waltham, MA) to 5 μl PCR product. The reaction mixture was put into a thermocycler with the following conditions: 37 °C for 15 min followed by 85 °C for 15 min. Sequencing of the reverse strand was done by Macrogen (Amsterdam, Netherlands).
Sequence processing and alignment were done using Bioedit (Hall 1999), the software package Geneious Version 10.2.6 (Biomatters Limited, Auckland, New Zealand) (Kearse et al. 2012), and the integrated MUSCLE algorithm (Edgar 2004). The received sequences were checked for contamination and clipped to a common length of 330 bp in order to obtain a complete alignment. Scheel et al. (2015) have published D-Loop sequences from additional samples of the same collection effort, which allowed for the inclusion of 48 additional sequences in all analyses (NCBI accession numbers: KP793607 - KP793669), creating a final alignment containing 122 samples (see Supplementary Information Table S1 for a complete list of samples).
Clustering of haplotypes
Haplotypes were inferred and a median-joining haplotype network (Bandelt et al. 1999) with ε = 0 was created using PopArt (Leigh and Bryant 2015). We then wanted to measure whether the geographic distance between capture sites among individuals with the same haplotype differed between sexes, thus indicating different degrees of clustering. For this, distances between all individuals of the same habitat and sex were calculated with Microsoft Excel using the trapping coordinates. We then calculated the mean distance between all individuals per habitat. Then, for each individual, the mean distance between the individual in question and individuals with the same haplotype occurring in the same habitat was calculated. This measure was called “Individual_Mean.” If haplotypes were distributed at random, the Individual_Mean would not differ from the mean distance between all individuals per habitat. If individuals with the same haplotype would be spatially clumped, the Individual_Mean would be smaller than the mean for all individuals; if individuals with the same haplotype were overdispersed, the Individual_Mean would be larger.
Microcebus griseorufus occurred in different densities in the three habitats, and thus, the distance measures differed between habitats due to the different densities. If there were the postulated differences in the spatial arrangement of haplotypes between habitats and we would use the “raw” means, we would not be able to distinguish between the effect of animal densities and the effect of habitat differences. Therefore, we calculated a standardized index for clustering, named Individual_Distance_by_Grand_Mean (ID). For this, we divided each individual mean distance by the mean distance per habitat. This accounted for habitat-specific differences in lemur densities. Values below 1 indicate lower distance than average (=clustering) and values above 1 indicate overdispersion. Graphics were created and statistical analyses were performed using Microsoft Excel and SPSS (Field 2013). Confidence intervals (95%) for medians were calculated using the bootstrap procedure with 10,000 runs of the Statistics Calculators by Montgomery College (https://pressbooks.montgomerycollege.edu/statcalcs/chapter/bootstrap-confidence-intervals/).
Haplotype group size
While the analysis of clustering provides a measure of spatial cohesiveness between individuals with the same haplotype, we use the number of individuals per haplotype as an indication of the number of individuals in population nuclei. Since we recorded the positions of animals during their active phase, the resulting measure of “group size” is not meant as a measure of the size of sleeping associations. Rather, it should represent the size of a group that, in the case of females, jointly defends important food resources that provide food reliably (sensu Génin 2008, 2010; Génin and Rambeloarivony 2018).
Results
Haplotype diversity
Using PopArt, we inferred 24 different mtDNA haplotypes from the sequence alignment containing 122 samples/sequences (Fig. 2, see Supplementary Information Table S2 for haplotype sequences). Among the 64 males, all 24 mtDNA haplotypes were represented, while only 12 of the haplotypes were found among the 58 females. The number of individuals per haplotype was higher in females than in males (Mann-Whitney U test; U = 167, nfemale = 15, nmale = 36, p = 0.034). All rare haplotypes (N=6), only represented by one individual, belonged to male mouse lemurs. The spatial arrangement of capture sites of the 122 sequenced mouse lemurs is illustrated in Fig. 3 for all three habitats.
Spatial distribution
Plotting the exact trapping coordinates for each vegetation type and sex shows clear differences in spatial distribution between the sexes (Fig. 4). While females show a clear pattern of haplotype clustering in Habitats 1 and 2 and less so in Habitat 3, males exhibit a more heterogeneous distribution of haplotypes, indicating increased dispersal. Female mouse lemurs inhabiting the dry forest on sandy soil (Habitat 1) clustered into five different haplotypes representing a total of 26 individuals, with two large central clusters containing 11 and 9 individuals, respectively. Males inhabiting Habitat 1 exhibited a total of 14 different haplotypes, represented by 26 individuals. Mouse lemurs inhabiting the dry forest on ferruginous soil (Habitat 2) exhibit a similar pattern. Female individuals in this habitat exhibited four different mtDNA haplotypes, but nearly all of them (N=15/17) belonged to only one of two haplotype clusters. The other two (haplotypes 5 and 17) were represented by single individuals. The 22 males observed here exhibited a total of 13 different haplotypes, which were heterogeneously dispersed across the whole transect. In the driest habitat, the xerophytic bush on calcareous soil (Habitat 3), females exhibit less clustering, constituting six different haplotypes represented by 15 individuals, with the two largest clusters representing five and four individuals, respectively. In the same habitat, nine mtDNA haplotypes were represented by 16 males.
Clustering
In order to compare the clustering of mouse lemurs between sexes and among habitats, individual aggregation was characterized by the Individual_Distance_by_Grand_Mean (ID; Fig. 5). Values below 1 indicate increased clustering while values above 1 indicate increased overdispersion. In all three habitats, females exhibited median ID values below 0.7; however, their significance differed according to habitat type. In the two forest habitats on sand (Habitat 1 and Habitat 2), the 95% confidence intervals of the median did not include 1, indicating significant clustering of haplotypes for females (C95% of the median, lower and upper limits: females: Habitat 1: 0.54–0.69; Habitat 2: 0.43–0.60). The lower and upper limits of the 95% confidence interval of the ID for females in Habitat 3 were 0.13–1.26 and thus indicated no consistent clustering of females in Habitat 3 (xerophytic bush) (Fig. 5). The median ID of females did not differ among the three habitats (Kruskal-Wallis analysis of variance: H = 1.894, df = 2, p = 0.388).
Clustering of female and male haplotypes by individual_distance_by_grand_mean (ID) value, comparing all three vegetation types across both sexes. Values are medians and quartiles; whiskers indicate the 1.5*interquartile range, outliers (circles) are outside the 3rd quartile + 1.5*interquartile range, extreme values (asterisks) are outside the 3rd quartile + 3*interquartile range
The 95% confidence intervals for male IDs in Habitat 1 and Habitat 2 include 1 and thus indicate no difference from randomness, with a tendency to being overdispersed, i.e., ID values above 1 (C95% of the median, lower and upper limits: males: Habitat 1: 1.00–1.13; Habitat 2: 0.97–1.15). In Habitat 3, the C95% of the median ID of male mouse lemurs also include 1 but exhibit a tendency towards clustering with a median ID of 0.86 (C95% 0.57–1.00). The median IDs of males do not differ between the three habitats (Kruskal-Wallis analysis of variance: H = 4.843, df = 2, p = 0.089).
The degree of clustering differs between females and males in Habitat 1 and Habitat 2 (Mann-Whitney U tests: Habitat 1: U = 624, p < 0.001; Habitat 2: U = 319, p < 0.001). The medians do not differ in Habitat 3 (Mann-Whitney U test: U = 138, p = 0.474). Thus, the distribution pattern (clustering) of females and males is distinct in the forests but converges towards randomness in the xerophytic bush (Fig. 5).
Haplotype group size
In females, the clusters of individuals belonging to the same haplotype in Habitat 1 and Habitat 2 consist of more individuals than in Habitat 3, while the number of individuals per haplotype remains constant in males (Fig. 6). Yet, since the actual number of haplotypes per habitat is small, neither the number of females nor the number of males per haplotype differs significantly among habitats (Kruskal-Wallis analysis of variance: females: H = 1.47; males: H = 0.16, df = 2, p > 0.4 in both tests).
Discussion
Overall, we recovered 24 different mtDNA haplotypes among 122 individuals, which shows a substantial level of genetic diversity in the mitochondrial DNA compared with other species. Wimmer et al. (2002) found 13 mtDNA haplotypes in 85 individuals of M. murinus, Kappeler et al. (2002) revealed 4 haplotypes among 46 individuals (sampled in 1 year) of the sympatric Mirza coquereli, and Gerloff et al. (1999) detected 5 haplotypes in a population of 36 bonobos. The mtDNA haplotype diversity of M. griseorufus (0.921), calculated according to Saitoh (2021), is higher than that of M. murinus (0.508) and M. coquereli (0.660). While it is difficult to compare different higher taxa, social units, and sampling areas, M. grisoeurufus is very diverse and ranks among the top 15% of the 66 terrestrial mammal species compiled by Saitoh (2021).
Genetic structure and social organization of M. griseorufus
The results of this study revealed that the genetic structure of Microcebus griseorufus is characterized by spatial clustering of females with shared haplotypes and dispersed males. This study detected a pronounced sex difference in the haplotype distribution of M. griseorufus. The high general diversity in haplotypes is mostly driven by males, which represent the majority of unique and rare haplotypes. Females (within a population) express fewer haplotypes that are generally shared with other females in proximity. There is a significant difference in the number of individuals per haplotype between males and females. This genetic structure is consistent with the results on M. griseorufus in Berenty where the population consists of population nuclei where related females live in close proximity and defend resources that provide food reliably (Génin 2008, 2010; Génin and Rambeloarivony 2018) and the findings on M. murinus with small family units of closely related females (Radespiel et al. 2001; Wimmer et al. 2002) and female philopatry, while high haplotype diversity and lack of spatial clustering of males imply dispersal. Males might have immigrated from other matrilineal clusters outside the study area. Since within subpopulations there are males sharing the most common haplotypes in females, it is possible that not all males disperse immediately after weaning. Alternatively, sons from distant female clusters with the same haplotype could have moved into the vicinity of females with the same haplotype. In other studies, this genetic organization has also been linked to or derived from sleeping associations (Radespiel et al. 2001; Wimmer et al. 2002; Rode et al. 2013). This might be an alternative correlation as there are very few large trees that would allow the formation of large sleeping associations (Table 1). But M. griseorufus seeks shelter in bundles of spiny branches that could compensate for the lack of hollows in large trees. While we did not analyze the genetics of sleeping associations, observations from Microcebus ganzhorni provide evidence that sleeping associations have different dynamics than the spatial associations of individuals during their nightly activity time (Lahann 2008). Since our study relied on captures of active individuals and did not reconstruct home ranges or monitored sleeping associations, we cannot resolve this issue.
Patterns of kinship are often invoked to explain the spatial aggregation of females in combination with affiliative and cooperative behavior to increase inclusive fitness (Sherman 1977; Gouzoules 1984; Packer et al. 1991; Moore 1992). The degree of kinship impacts cooperative behavior in anthropoid primates (Morin et al. 1994; Pope 2000; Chapais 2001; Silk 2002), though cooperation between unrelated individuals has also been reported for bonobos and chimpanzees (Goldberg and Wrangham 1997; Gerloff et al. 1999; Mitani et al. 2000). In M. griseorufus, the evolutionary benefit of forming groups of related females could be a better defense of rewarding food resources that result in higher reproductive success of kin (Génin and Rambeloarivony 2018), as well as mutual nursing of offspring from related females (Eberle and Kappeler 2006). Alternatively, females avoid the risks of dispersal and increase thus their fitness even without any benefits from defending resources.
These findings on the social organization of M. griseorufus are in line with the research of the well-studied Microcebus murinus, which exhibits female sleeping associations and dispersed males (Radespiel et al. 2001, 2003; Wimmer et al. 2002; Fredsted et al. 2004, 2005). Microcebus murinus, at the center of its distribution range, shows a similar genetic structure as M. griseorufus, living at the dry environmental limits of the genus (Bohr et al. 2011; Steffens et al. 2017). This study of M. griseorufus might hint towards the unity of social organizations, across a large environmental gradient. This is consistent with behavioral observations of different species of Microcebus from all major forest types in Madagascar. In all species, males have larger home ranges than females (M. murinus: Fietz 1999; M. rufus: Atsalis 2000; M. berthae: Schwab 2000; Schwab and Ganzhorn 2004; Dammhahn and Kappeler 2005; M. ganzhorni: Lahann 2008). But this is where comparative data come to their limits. The composition of sleeping groups has been used to interpret the social organization in a similar way as the spatial clusters of related females. Data are available for M. rufus (Karanewsky and Wright 2015), M. berthae (Schwab 2000; Dammhahn and Kappeler 2005), M. griseorufus (Génin 2008), M. ganzhorni (Lahann 2008), M.ravelobensis (Radespiel et al. 2009), and M. sambiranensis (Hending et al. 2017). Most of these studies described sleeping groups as highly dynamic with changing composition, sometimes involving members of both sexes. The analogy of sleeping groups and clusters of related females may not be wrong but sleeping groups are something different than the spatial clusters documented by animals during their active phase. Though some sleeping groups allow for alloparental care (Eberle and Kappeler 2006; Génin 2008), the evolutionary relevance of sleeping groups is unclear.
It is not uncommon for members of a genus to share a social organization. In Madagascar, all nine species of wooly lemurs (Avahi) live in pairs, and all nine species of the genus Propithecus are organized in groups (Donati et al. 2022; Lawler and Richard 2022). However, there are also studies indicating a range of different social organizations and social systems within the same species or genus. Varecia shows substantial variation in grouping patterns (Vasey et al. 2022), and most Eulemur species share a social organization across a variety of habitats, but Eulemur rubriventer and Eulemur mongoz form pair-bonded family groups while the other Eulemur species live in larger multi-male multi-female groups (Johnson et al., 2022). Baboons of the genus Papio comprise six closely related species that show ecological plasticity and different social organizations. Four species of baboons exhibit uni-level organization with big groups consisting of multiple males and females, while two other species display multi-level social organizations with one-male-units, parties, and gangs (Barton et al. 1996; Fischer et al. 2019; Zinner et al. 2021).
Plasticity of the social organization along an ecological gradient
The socioecological model predicts female spatial distribution according to ecological factors such as resource availability and risk avoidance (e.g., predation pressure, dispersal risks, disease transmission). The availability of receptive females, in contrast, affects the spatial distribution of males trying to maximize their access to said females. Analyzing M. griseorufus across their natural habitat range in three distinct vegetation types offers the advantage of a pronounced ecological gradient at a small local scale. The habitat data published by Ratovonamana et al. (2011) show a decline in the density of food plants as well as fewer large trees (with tree holes as shelters) available in the xerophytic bush compared to the forest habitats. Assuming that other ecological factors such as predation pressure and risk of dispersal should be similar because of the close proximity of the different habitats, changes in their social organization should largely reflect a response to resource availability. The capture data in these habitats show a corresponding abundance of mouse lemurs to resource availability.
The general social organization of female philopatry with dispersed males is found across all three habitats regardless of the ecological conditions. These results are in line with other mammalian studies providing evidence of fixed social systems in genera or species across different ecological conditions. Even though it was hypothesized that the social organization of hamadryas baboons is an adaptation to the harsh ecological conditions they are facing, Guinea baboons exhibit the same multi-level social organization, despite mostly living in vastly different habitats (Zinner et al. 2021). Thus, some adaptivity in social characteristics might be explained by phylogeny rather than by ecology (Thierry et al. 2000; Menard 2004; Ossi and Kamilar 2006; Thierry 2007; Shultz et al. 2011; Schradin 2013). There are studies finding contrary evidence, however, showing plasticity in social organization as a hypothesized response to ecological change. Koenig et al. (1998) found differences in female social relationships in Hanuman langurs (Semnopithecus entellus) between populations as an assumed adaptation to ecological conditions. This is paralleled in Propithecus diadema, Eulemur collaris, and other lemur species where group size and group cohesion declined within populations with increasing fragmentation and degradation of forests and reduced food patch sizes (Ganzhorn 1988; Irwin 2007; Donati et al. 2011). Eulemur exhibits a variety of behaviors in response to their environment across the genus (Kappeler and Fichtel 2016), while the social organization is more related to phylogenetic distance between species (Ossi and Kamilar 2006). Izar et al. (2012) found that social relationships between female tufted capuchin monkeys (Sapajus) changed between two species in accordance with their food sources, while the mating system seemed to be constrained by phylogenetic inertia. Similarly, macaques (Macaca), as the most geographically widespread primate taxon show the same grouping and dispersal patterns across their 22 species, while displaying unparalleled diversity in behavior and relationships (Thierry et al. 2000; Thierry 2007).
It is to be noted that while the clustering of females with shared haplotypes in M. griseorufus does not significantly differ among habitats, female aggregation is lower in the xerophytic bush and is not significantly different from a random distribution. This suggests that when resources are not defendable or not worth defending, female associations might decrease. At the same time, males tend to be overdispersed in the forest habitats, indicating competition among individuals. While the distribution of males does not differ significantly from randomness according to our measure, their organization changes drastically between forests and the xerophytic shrub (Habitat 3). It might be possible, that the genetically determined social organization of M. griseorufus starts to crumble at the dry edge of their tolerance. The reduced density of mouse lemurs inhabiting the xerophytic shrub is probably a response to the scarcity of food and nesting sites. The spatial clustering of females is more pronounced in the forest ecosystems, with higher availability of food and trees. This pattern matches the predictions of the socioecological model that anticipate a decline in group size with the declining size of available food patches.
While our findings are coherent with Thierry’s and Clutton-Brock and Janson’s assessment of the socioecological model (Thierry 2008; Clutton-Brock and Janson 2012), we cannot rule out the possibility that M. griseorufus has different population densities in the three habitats that are unrelated to resource availabilities. If so, the observed differences in social organization could be a consequence of differing population densities. Changes of this kind have been reviewed by Schradin et al. (2020). By using several populations of African striped mice with different constellations of resource abundance and population densities, they could show that changes in the social organization were density-dependent but unrelated to food resources. We tried to account for different population densities by introducing the measure of “Individual_Distance_by Grand_Mean” that represents a standardization that accounts for the different population densities. Yet, this option cannot be ruled out.
Some primate behavior, such as feeding competition, is undoubtedly influenced by their ecological surroundings. Nevertheless, other traits might be better explained by phylogeny. This is exemplified by Guinea and hamadryas baboons that live under very different ambient conditions but show very similar social organizations (Fischer et al. 2019; Zinner et al. 2021). It is also to be considered that primate societies might not be optimal (Janson 2000), and the number of social organizations that are possible is underlain by stabilizing selection preventing adaptations needed for better survival in any given environment (Thierry 1997, 2007, 2008; Hemelrijk 2002). This would allow for some diversity in social organization (Di Fiore and Rendall 1994) while restricting adaptations to ecological characteristics (Fleagle and Reed 1996; Thierry 2008).
The present study offers a perspective on how the social organization of a mouse lemur can withstand the ecological pressures of degrading habitats to a certain extent, but eventually, the genetic structure seems to collapse. The consequences of a collapse of the social organization are hard to predict for different species of this genus. Cooperative breeding and nursing offspring of related females improves the reproductive success of M. murinus and M. griseorufus (Eberle and Kappeler 2006; Génin 2008). This option is probably reduced at the dry limits of the species ranges. But since our data are based on year-round captures and did not allow for a finer temporal resolution, we could not exclude the option of seasonal affiliations for raising offspring. At least, M. murinus seems to be able to compensate for environmental changes by accelerating their life histories (Ozgul et al. 2023). Other Microcebus species, such as the sympatric M. berthae, do not seem to have this plasticity and seem to be on their way to extinction (Kappeler et al. 2022). (In conclusion, the social organization of the gray-brown mouse lemur, Microcebus griseorufus, can be characterized by spatial clusters of females sharing the same haplotype while male haplotypes are overdispersed.) The principle of this structure persists across an environmental gradient towards the environmental limit for the species, but the number of individuals per cluster with the same haplotype changes in a similar way as group size is predicted to change by the socioecological model. At the dry limit of their tolerance, when less food and nesting sites are available, this structure seems to collapse, and the spatial arrangement of haplotypes does not deviate from random. For the time being, the consequences of these changes in the social organization are not known.
Data availability
The sequence data used in this study are available in the NCBI GenBank with the accession numbers KP793607 - KP793669 and OQ605019 - OQ605092. All other data supporting the findings of the study can be found in this article and the supplementary information files.
References
Andriatsimietry R, Goodman SM, Razafimahatratra E, Jeglinski JWE, Marquard M, Ganzhorn JU (2009) Seasonal variation in the diet of Galidictis grandidieri Wozencraft, 1986 (Carnivora: Eupleridae) in a sub-arid zone of extreme south-western Madagascar. J Zool 279:410–415
Atsalis S (2000) Spatial distribution and population composition of the brown mouse lemur (Microcebus rufus) in Ranomafana National Park, Madagascar, and its implications for social organization. Am J Primatol 51:61–78
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Barton RA, Byrne RW, Whiten A (1996) Ecology, feeding competition and social structure in baboons. Behav Ecol Sociobiol 38:321–329
Bohr YE-MB, Giertz P, Ratovonamana YR, Ganzhorn JU (2011) Gray-brown mouse lemurs (Microcebus griseorufus) as an example of distributional constraints through increasing desertification. Int J Primatol 32:901–913
Brockman HJ (2001) The evolution of alternative strategies and tactics. Adv Stud Behav 30:1–51
Cassini MH (2021) Sexual aggression in mammals. Mammal Rev 51:247–255
Chapais B (2001) Primate nepotism: what is the explanatory value of kin selection? Int J Primatol 22:203–229
Chapman CA, Balcomb SR, Gillespie TR, Skorupa JP, Strusaker TT (2000) Long-term effects of logging on African primate communities: a 28-year comparison from Kibale National Park, Uganda. Conserv Biol 14:207–217
Clutton-Brock T, Janson C (2012) Primate socioecology at the crossroads: past, present, and future. Evol Anthropol 21:136–150. https://doi.org/10.1002/evan.21316
Crook JH (1964) The evolution of social organisation and visual communication in the weaver birds (Ploceinae). Behaviour Suppl 10:1–178
Dammhahn M, Kappeler PM (2005) Social system of Microcebus berthae, the World’s smallest primate. Int J Primatol 26:407–435. https://doi.org/10.1007/s10764-005-2931-z
Dewar RE, Richard AF (2007) Evolution in the hypervariable environment of Madagascar. P Natl Acad Sci USA 104:13723–13727
Di Fiore A, Rendall D (1994) Evolution of social organization: a reappraisal for primates by using phylogenetic methods. P Natl Acad Sci USA 91:9941–9945
Donati G, Kesch K, Ndremifidy K, Schmidt SL, Ramanamanjato JB, Borgognini-Tarli SM, Ganzhorn JU (2011) Better few than hungry: flexible feeding ecology of collared lemurs Eulemur collaris in littoral forest fragments. PLoS ONE 6:e19807
Donati G, Norscia I, Balestri M, Zaramody A, Louis EE Jr, Thalmann U (2022) Indriidae: Avahi, Woolly Lemurs, Fotsy-fe, Tsarafangitra, Dadintsifaky. In: Goodman SM (ed) The new natural history of Madagascar. Princeton University Press, Princeton, pp 1963–1967
Dunbar RIM (1988) Primate social systems. Springer, New York
Eberle M, Kappeler PM (2006) Family insurance: kin selection and cooperative breeding in a solitary primate (Microcebus murinus). Behav Ecol Sociobiol 60:582–588
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Emlen ST, Oring LW (1977) Ecology, sexual selection, and the evolution of mating systems. Science 197:215–223
Estrada A, Garber PA, Rylands AB et al (2017) Impending extinction crisis of the world’s primates: Why primates matter. Sci Adv 3:e1600946
Field A (2013) Discovering statistics using IBM SPSS Statistics. SAGE, London
Fietz J (1999) Mating system of Microcebus murinus. Am J Primatol 48:127–133
Fischer J, Higham JP, Alberts SC et al (2019) Insights into the evolution of social systems and species from baboon studies. Elife 8:e50989
Fleagle JG, Reed KE (1996) Comparing primate communities: a multivariate approach. J Hum Evol 30:489–510
Fredsted T, Pertoldi C, Olesen JM, Eberle M, Kappeler PM (2004) Microgeographic heterogeneity in spatial distribution and mtDNA variability of gray mouse lemurs (Microcebus murinus, Primates: Cheirogaleidae). Behav Ecol Sociobiol 56:393–403. https://doi.org/10.1007/s00265-004-0790-9
Fredsted T, Pertoldi C, Schierup MH, Kappeler PM (2005) Microsatellite analyses reveal fine-scale genetic structure in grey mouse lemurs (Microcebus murinus). Mol Ecol 14:2363–2372. https://doi.org/10.1111/j.1365-294X.2005.02596.x
Ganzhorn JU (1988) Food partitioning among Malagasy primates. Oecologia 75:436–450
Génin F (2008) Life in unpredictable environments: first investigation of the natural history of Microcebus griseorufus. Int J Primatol 29:303–321. https://doi.org/10.1007/s10764-008-9243-z
Génin F (2010) Who sleeps with whom? Sleeping association and socio-territoriality in Microcebus griseorufus. J Mammal 91:942–951. https://doi.org/10.1644/09-MAMM-A-239.1
Génin F, Rambeloarivony H (2018) Mouse lemurs (Primates: Cheirogaleidae) cultivate green fruit gardens. Biol J Linn Soc 124:607–620. https://doi.org/10.1093/biolinnean/bly087
Gerloff U, Hartung B, Fruth B, Hohmann G, Tautz D (1999) Intracommunity relationships, dispersal pattern and paternity success in a wild living community of Bonobos (Pan paniscus) determined from DNA analysis of faecal samples. Proc R Soc Lond B 266:1189–1195. https://doi.org/10.1098/rspb.1999.0762
Goldberg TL, Wrangham RW (1997) Genetic correlates of social behaviour in wild chimpanzees: evidence from mitochondrial DNA. Anim Behav 54:559–570
Goldizen AW (1990) A comparative perspective on the evolution of tamarin and marmoset social systems. Int J Primatol 11:63–83
Gouzoules S (1984) Primate mating systems, kin associations, and cooperative behavior: evidence for kin recognition? Am J Phys Anthropol 27:99–134
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid S 41:95–98
Hamilton WD (1964) The genetical evolution of social behaviour. II. J Theor Biol 7:17–52
Hannah L, Dave R, Lowry PP, Andelman S, Andrianarisata M, Andriamaro L, Cameron A, Hijmans R, Kremen C, MacKinnon J (2008) Climate change adaptation for conservation in Madagascar. Biol Lett 4:590–594
Hapke A, Gligor M, Rakotondranary SJ, Rosenkranz D, Zupke O (2011) Hybridization of mouse lemurs: different patterns under different ecological conditions. BMC Evol Biol 11:297. https://doi.org/10.1186/1471-2148-11-297
Hemelrijk CK (2002) Understanding social behaviour with the help of complexity science. Ethology 108:655–671. https://doi.org/10.1046/j.1439-0310.2002.00812.x
Hending D, McCabe G, Holderied M (2017) Sleeping and ranging behavior of the Sambirano Mouse Lemur, Microcebus sambiranensis. Int J Primatol 38:1072–1089
Ims RA (1988) The potential for sexual selection in males: effect of sex ratio and spatiotemporal distribution of receptive females. Evol Ecol 2:338–352
Irwin MT (2007) Living in forest fragments reduces group cohesion in diademed sifakas (Propithecus diadema) in eastern Madagascar by reducing food patch size. Am J Primatol 69:434–447
Irwin MT, Wright PC, Birkinshaw C et al (2010) Patterns of species change in anthropogenically disturbed forests of Madagascar. Biol Conserv 143:2351–2362
Izar P, Verderane MP, Peternelli-dos-Santos L, Mendonça-Furtado O, Presotto A, Tokuda M, Visalberghi E, Fragaszy D (2012) Flexible and conservative features of social systems in tufted capuchin monkeys: comparing the socioecology of Sapajus libidinosus and Sapajus nigritus. Am J Primatol 74:315–331. https://doi.org/10.1002/ajp.20968
Janson CH (1992) Evolutionary ecology of primate social structure. In: Smith EA, Winterhalder B (eds) Evolutionary ecology and human behavior. Routledge, London
Janson CH (2000) Primate socio-ecology: the end of a golden age. Evol Anthropol 9:73–86
Johns AD, Skorupa JP (1987) Responses of rain-forest primates to habitat disturbance: a review. Int J Primatol 8:157–191
Johnson SE, Tecot SR, Ralainasolo FB, Ratsimbazafy JH, Overdorff DJ, Donati G (2022) Lemuridae: eulemur, true lemurs. In: Goodman SM (ed) The new natural history of Madagascar. Princeton University Press, Princeton, pp 1941–1947
Kappeler PM, Fichtel C (2016) The evolution of Eulemur social organization. Int J Primatol 37:10–28
Kappeler PM, Radespiel U, Rasoloarison MR, Salmona J, Yoder AD (2022) Cheirogaleidae: microcebus, mouse lemurs, tsidy, tsy-tsy. In: Goodman SM (ed) The new natural history of Madagascar. Princeton University Press, Princeton, pp 1927–1932
Kappeler PM, van Schaik CP (2002) Evolution of primate social systems. Int J Primatol 23:707–740. https://doi.org/10.1023/A:1015520830318
Kappeler PM, Wimmer B, Zinner D, Tautz D (2002) The hidden matrilineal structure of a solitary lemur: implications for primate social evolution. Proc R Soc Lond B 269:1755–1763. https://doi.org/10.1098/rspb.2002.2066
Karanewsky CJ, Wright PC (2015) A preliminary investigation of sleeping site selection and sharing by the brown mouse lemur Microcebus rufus during the dry season. J Mammal 96:1344–1351
Kasola C, Atrefony F, Louis F, Odilon GN, Ralahinirina RG, Menjanahary T, Ratovonamana YR (2020) Population dynamics of Lemur catta at selected sleeping sites in the Tsimanampesotse National Park. Malagasy Nature 14:69–80
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Koenig A, Beise J, Chalise MK, Ganzhorn JU (1998) When females should contest for food – testing hypotheses about resource density, distribution, size, and quality with Hanuman langurs (Presbytis entellus). Behav Ecol Sociobiol 42:225–237. https://doi.org/10.1007/s002650050434
Koenig A, Borries C (2009) The lost dream of ecological determinism: time to say goodbye?… or a white queen's proposal? Evol Anthropol 18:166–174
Koenig A, Scarry CJ, Wheeler BC, Borries C (2013) Variation in grouping patterns, mating systems and social structure: what socio-ecological models attempt to explain. Phil Trans R Soc B 368:20120348. https://doi.org/10.1098/rstb.2012.0348
Krebs JR, Davies NB (1978) Behavioural ecology - an evolutionary approach. Blackwell Scientific Publications, Oxford
Lahann P (2008) Habitat utilization of three sympatric cheirogaleid lemur species in a littoral rain forest of southeastern Madagascar. Int J Primatol 29:117–134
Lawler RR, Richard AF (2022) Indriidae: Propithecus, Sifakas, Sifaka. In: Goodman SM (ed) The new natural history of Madagascar. Princeton University Press, Princeton, pp 1971–1975
Leigh JW, Bryant D (2015) popart: full-feature software for haplotype network construction. Meth Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
Lott DF (1984) Intraspecific variation in the social systems of wild vertebrates. Behaviour 88:266–325
Mamokatra A (1999) Etude pour l’élaboration d’un plan d’aménagement et de gestion au niveau de la Réserve Naturelle Intégrale de Tsimanampetsotsa. In: Diagnostic physico-bio-écologique. Deutsche Forstservice GmbH, Feldkirchen et Entreprise d’Etudes de Développement Rural “Mamokatra”, Antananarivo, Madagascar
Menard N (2004) Do ecological factors explain variation in social organization? In: Thierry B, Singh M, Kaumanns W (eds) Macaque societies: a model for the study of social organization. Cambridge University Press, Cambridge, pp 237–261
Mitani JC, Merriwether DA, Zhang C (2000) Male affiliation, cooperation and kinship in wild chimpanzees. Anim Behav 59:885–893
Mittermeier RA, Louis EE Jr, Richardson MJ, Schwitzer C, Langrand O, Rylands AB, Hawkins F, Rajaobelina S, Ratsimbazafy J, Rasoloarison R (2010) Lemurs of madagascar. Conservation International, Arlington
Moat J, Smith PP (2007) Atlas of the vegetation of Madagascar. Royal Botanic Gardens, Kew
Moore J (1992) Dispersal, nepotism, and primate social behavior. Int J Primatol 13:361–378
Morin PA, Moore JJ, Chakraborty R, Jin L, Goodall J, Woodruff DS (1994) Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science 265:1193–1201
Ossi K, Kamilar JM (2006) Environmental and phylogenetic correlates of eulemur behavior and ecology (Primates: Lemuridae). Behav Ecol Sociobiol 61:53–64. https://doi.org/10.1007/s00265-006-0236-7
Ozgul A, Fichtel C, Paniw M, Kappeler PM (2023) Destabilising effect of climate change on the persistence of a short-lived primate. P Natl Acad Sci USA 120:e2214244120. https://doi.org/10.1073/pnas.2214244120
Packer C, Gilbert DA, Pusey AE, O’Brieni SJ (1991) A molecular genetic analysis of kinship and cooperation in African lions. Nature 351:562–565
Peres CA, Gardner TA, Barlow J, Zuanon J, Michalski F, Lees AC, Vieira ICG, Moreira FMS, Feeley KJ (2010) Biodiversity conservation in human-modified Amazonian forest landscapes. Biol Conserv 143:2314–2327
Pope TR (2000) Reproductive success increases with degree of kinship in cooperative coalitions of female red howler monkeys (Alouatta seniculus). Behav Ecol Sociobiol 48:253–267
Pulliam HR, Caraco T (1984) Living in groups: is there an optimal group size? In: Krebs JR, Davies NB (eds) Behavioural ecology - an evolutionary approach. Blackwell Scientific Publications, Oxford, pp 122–147
Radespiel U, Cepok S, Zietemann V, Zimmermann E (1998) Sex-specific usage patterns of sleeping sites in grey mouse lemurs (Microcebus murinus) in northwestern Madagascar. Am J Primatol 46:77–84
Radespiel U, Juric M, Zimmermann E (2009) Sociogenetic structures, dispersal and the risk of inbreeding in a small nocturnal lemur, the golden-brown mouse lemur (Microcebus ravelobensis). Behaviour 146:607–628
Radespiel U, Lutermann H, Schmelting B, Bruford MW, Zimmermann E (2003) Patterns and dynamics of sex-biased dispersal in a nocturnal primate, the grey mouse lemur, Microcebus murinus. Anim Behav 65:709–719. https://doi.org/10.1006/anbe.2003.2121
Radespiel U, Lutermann H, Schmelting B, Zimmermann E (2019) An empirical estimate of the generation time of mouse lemurs. Am J Primatol 81:e23062
Radespiel U, Sarikaya Z, Zimmermann E, Bruford MW (2001) Sociogenetic structure in a free-living nocturnal primate population: sex-specific differences in the grey mouse lemur (Microcebus murinus). Behav Ecol Sociobiol 50:493–502
Rakotondranary JS, Ratovonamana YR, Ganzhorn JU (2010) Distributions et caractéristiques des microhabitats de Microcebus griseorufus (Cheirogaleidae) dans le Parc National de Tsimanampetsotsa (Sud-ouest de Madagascar). Malagasy Nature 4:55–64
Rasoazanabary E (2004) A preliminary study of mouse lemurs in the Beza Mahafaly Special Reserve, southwest Madagascar. Lemur News 9:4–7
Ratovonamana RY, Rajeriarison C, Roger E, Ganzhorn JU (2011) Phenology of different vegetation types in Tsimanampetsotsa National Park, southwestern Madagascar. Malagasy Nature 5:14–38
Rode EJ, Nekaris KAI, Markolf M, Schliehe-Diecks S, Seiler M, Radespiel U, Schwitzer C (2013) Social organisation of the northern giant mouse lemur Mirza zaza in Sahamalaza, north western Madagascar, inferred from nest group composition and genetic relatedness. Contrib Zool 82:71–83
Ross KG (2001) Molecular ecology of social behaviour: analyses of breeding systems and genetic structure. Mol Ecol 10:265–284
Saitoh T (2021) High variation of mitochondrial DNA diversity as compared to nuclear microsatellites in mammalian populations. Ecol Res 36:206–220. https://doi.org/10.1111/1440-1703.12190
Scheel BM, Henke-von der Malsburg J, Giertz P, Rakotondranary SJ, Hausdorf B, Ganzhorn JU (2015) Testing the influence of habitat structure and geographic distance on the genetic differentiation of mouse lemurs (Microcebus) in Madagascar. Int J Primatol 36:823–838. https://doi.org/10.1007/s10764-015-9855-z
Schliehe-Diecks S, Eberle M, Kappeler PM (2012) Walk the line—dispersal movements of gray mouse lemurs (Microcebus murinus). Behav Ecol Sociobiol 66:1175–1185
Schradin C (2013) Intraspecific variation in social organization by genetic variation, developmental plasticity, social flexibility or entirely extrinsic factors. Phil Trans R Soc B 368:20120346
Schradin C, Drouard F, Lemonnier G, Askew R, Olivier CA, Pillay N (2020) Geographic intra-specific variation in social organization is driven by population density. Behav Ecol Sociobiol 74:113
Schradin C, Hayes LD, Pillay N, Bertelsmeier C (2018) The evolution of intraspecific variation in social organization. Ethology 124:527–536
Schwab D (2000) A preliminary study of spatial distribution and mating system of pygmy mouse lemurs (Microcebus cf myoxinus). Am J Primatol 51:41–60
Schwab D, Ganzhorn JU (2004) Distribution, population structure and habitat use of Microcebus berthae compared to those of other sympatric cheirogalids. Int J Primatol 25:307–330. https://doi.org/10.1023/B:IJOP.0000019154.17401.90
Schwagmeyer PL (1988) Scramble-competition polygyny in an asocial mammal: male mobility and mating success. Am Nat 131:885–892
Sherman PW (1977) Nepotism and the evolution of alarm calls: alarm calls of Belding’s ground squirrels warn relatives, and thus are expressions of nepotism. Science 197:1246–1253
Shultz S, Opie C, Atkinson QD (2011) Stepwise evolution of stable sociality in primates. Nature 479:219–222
Silk JB (2002) Kin selection in primate groups. Int J Primatol 23:849–875
Sodhi NS, Koh LP, Clements R, Wanger TC, Hill JK, Hamer KC, Clough Y, Tscharntke T, Posa MRC, Lee TM (2010) Conserving Southeast Asian forest biodiversity in human-modified landscapes. Biol Conserv 143:2375–2384
Steffens KJE, Rakotondranary JS, Ratovonamana YR, Ganzhorn JU (2017) Vegetation thresholds for the occurrence and dispersal of Microcebus griseorufus in southwestern Madagascar. Int J Primatol 38:1138–1153. https://doi.org/10.1007/s10764-017-0003-9
Steffens KJE, Sanamo J, Razafitsalama J, Ganzhorn JU (2023) Ground-based vegetation descriptions and remote sensing as complementary methods describing habitat requirements of a frugivorous primate in northern Madagascar: implications for forest restoration. Anim Conserv. https://doi.org/10.1111/acv.12839
Sterck EH, Watts DP, van Schaik CP (1997) The evolution of female social relationships in nonhuman primates. Behav Ecol Sociobiol 41:291–309
Strier KB (2017) What does variation in primate behavior mean? Am J Phys Anthropol 162:4–14
Tadross M, Randriamarolaza L, Rabefitia Z, Zheng KY (2008) Climate change in Madagascar; recent past and future. World Bank, Washington DC
Terborgh J, Janson CH (1986) The socioecology of primate groups. Annu Rev Ecol Syst 17:111–136. https://doi.org/10.1146/annurev.es.17.110186.000551
Thierry B (1997) Adaptation and self-organization in primate societies. Diogenes 45:39–71
Thierry B (2007) Unity in diversity: lessons from macaque societies. Evol Anthropol 16:224–238. https://doi.org/10.1002/evan.20147
Thierry B (2008) Primate socioecology, the lost dream of ecological determinism. Evol. Anthropol 17:93–96. https://doi.org/10.1002/evan.20168
Thierry B, Iwaniuk AN, Pellis SM (2000) The influence of phylogeny on the social behaviour of macaques (Primates: Cercopithecidae, genus Macaca). Ethology 106:713–728. https://doi.org/10.1046/j.1439-0310.2000.00583.x
Trivers RL (1972) Parental investment and sexual selection. In: Campbell B (ed) Sexual Selection and the descent of man, 1871-1971. Aldine, Chicago, pp 136–179
van Schaik CP (1989) The ecology of social relationships amongst female primates. In: Standen V, Foley RA (eds) Comparative socioecology. The behavioural ecology of humans and other mammals, Blackwell, Oxford, pp 195–218
van Schaik CP (1983) Why are diurnal primates living in groups? Behaviour 87:120–144
Vasey N, Baden AL, Ratsimbazafy JH (2022) Lemuridae: Varecia, ruffed lemurs, Varikandana, Varijatsy. In: Goodman SM (ed) The new natural history of Madagascar. Princeton University Press, Princeton, pp 1957–1963
Weidt A, Hagenah N, Randrianambinina B, Radespiel U, Zimmermann E (2004) Social organization of the golden brown mouse lemur (Microcebus ravelobensis). Am J Phys Anthropol 123:40–51
Wilson EO (1975) Sociobiology: the new synthesis. Harvard University Press, Cambridge
Wimmer B, Tautz D, Kappeler P (2002) The genetic population structure of the gray mouse lemur (Microcebus murinus), a basal primate from Madagascar. Behav Ecol Sociobiol 52:166–175. https://doi.org/10.1007/s00265-002-0497-8
Wrangham RW (1980) An ecological model of female-bonded primate groups. Behaviour 75:262–300. https://doi.org/10.1163/156853980X00447
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395–420
Zinner D, Klapproth M, Schell A, Ohrndorf L, Chala D, Ganzhorn JU, Fischer J (2021) Comparative ecology of Guinea baboons (Papio papio). Primate Biol 8:19–35. https://doi.org/10.5194/pb-8-19-2021
Acknowledgements
The study was carried out under the “Accord de Collaboration” between Madagascar National Parks (MNP), the University of Antananarivo, and the University of Hamburg. We acknowledge the authorization and support of this study by the Ministère de l’Environement, des Eaux et Forêts et du Tourisme, MNP, and the University of Antananarivo. We thank the Malagasy authorities for issuing the research permit. Fieldwork was authorized by the Ministère de L’Environnement, de l’Ecologie, de la Mer et des Forêts. We thank MNP, Chantal Andrianarivo, Jocelyn Rakotomalala, Domoina Rakotomalala, the late Olga Ramilijaona, Daniel Rakotondravony, Charlotte Rajeriarison, and Roger Edmond for their collaboration and support. Tolona Andrianasolo kindly managed administrative affairs. Kasola Charles (Edson), Fisy Luis, and Kateffona Florent (Antsara) provided important help in the field. For their excellent support, we acknowledge Sabine Baumann, Yvonne Bohr, Jutta Hammer, Matthias Marquart, Jana Jeglinski, Iris Kiefer, Susanne Kobbe, and the staff of the camp Andranovao. We also want to thank Irene Tomaschewski for her help with DNA extractions. Comments on the manuscript by Kevin Langergraber and two reviewers are highly appreciated.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was funded by the Deutsche Forschungsgemeinschaft (Ga 342/15), Volkswagen Foundation, and WWF Germany.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
All animal work was conducted according to Malagasy and German guidelines. Our research was conducted in collaboration with the Département Biologie Animale and the Département Biologie et Ecologie Végétale of the Université d’Antananarivo. Authorization to enter Tsimanampetsotsa National Park as well as to capture and handle small mammals were delivered by the Ministère de l’Environement, des Eaux et Forêts et du Tourisme of Madagascar in accordance of Madagascar National Parks (MNP, former ANGAP; permit n° 057/07 issued on March 12, 2007, permit n° 009/08 issued on January 15, 2008, and permit n° 261/08 issued on October 9, 2008). We hereby confirm that our study was conducted in accordance with the recommendations of the Weatherall report "The use of non-human primates in research". The research was approved by the Ethics commission of the Institute of Zoology of the Universität Hamburg.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by K. Langergraber
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(XLSX 21 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abel, C., Giertz, P., Ratovonamana, Y.R. et al. Habitat quality affects the social organization in mouse lemurs (Microcebus griseorufus). Behav Ecol Sociobiol 77, 65 (2023). https://doi.org/10.1007/s00265-023-03339-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00265-023-03339-1