Abstract
Purpose
The association between TNF-α-308(G/A) and -238(G/A) polymorphisms and the susceptibility of non-traumatic osteonecrosis of the femoral head (NONFH) was investigated in many studies with conflicting results. We aimed to conduct a meta-analysis to evaluate the relationship between them comprehensively.
Methods
Relevant literatures published in PubMed, Web of Science, Embase, Cochrane library databases, China National Knowledge Infrastructure (CNKI), WANFANG Data, and China Science and Technology Journal Database (CSTJ) updated to January 30, 2018, were reviewed by two investigators independently. Odds ratios (ORs) and its 95% confidence intervals (95% CIs) were calculated by a fixed-effect model based on the indistinctive heterogeneity.
Results
For TNF-α-308(G/A) polymorphism, we recruited five studies including 432 NONFH patients and 760 controls and a statistically significant association was identified in Asians in four modes consisting of alleles mode (OR = 0.648, 95% CI 0.475–0.885), homozygote mode (OR = 0.330, 95% CI 0.136–0.802), dominant mode (OR = 0.344, 95% CI 0.143–0.827), and recessive mode (OR = 0.674, 95% CI 0.468–0.971), but no significant association was observed in Caucasians. For TNF-α-238(G/A) polymorphism, three eligible studies including 275 cases and 610 controls were evaluated and there was a significant association in alleles mode (OR = 0.270, 95% CI 0.4148–0.490) as well as recessive mode (OR = 0.254, 95% CI 0.138–0.468).
Conclusion
This meta-analysis shows that TNF-α-308(G/A) and -238(G/A) polymorphisms are associated with the susceptibility of NONFH, while the significant association for 308(G/A) is mainly observed in Asians.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
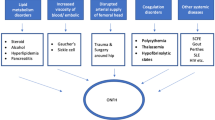
Osteonecrosis is a potentially devastating condition in which the reduction of blood supply is the most significant motivator [1,2,3]. When it comes to non-traumatic osteonecrosis of the femoral head (NONFH) which is a clinically common osteoarthropathy that mostly affects adults between 30 and 50 years old [4], the aetiology and pathology are not completely clarified [5]. Numerous risk factors have been identified that contribute to NONFH or the relevant pathological conditions, including hypercortisonism [6], hyperlipidemia, autoimmune diseases, dysbaric phenomena, smoking, hypofibrinolysis, alcoholism, and clotting disturbances [7, 8], which might result in intravascular coagulation and limited blood flow in caput femoris. Although plenty of risk factors have been recognized, the pathogenesis in molecular level remains unclear [9] and the therapy for osteonecrosis of femoral head continues to be a challenging.
Tumour necrosis factor (TNF), first reported in 1975 by Dr. Lloyd J. Old from Memorial Sloan-Kettering Cancer Center, New York, was described as a substance that mediates endotoxin-induced necrosis of tumors [10]. After years of investigation, TNF-α is generally known as a cell-signaling protein or cytokine participating in systemic inflammation and acute phase reaction. Produced mainly by macrophages as well as other immune effector cells and regulatory cells [11], TNF-α plays a rather intricate role in cellular processes: growth, proliferation, differentiation, and the immune reaction [12]. Meanwhile, the accommodative dysfunction of TNF-α has been reported to be implicated with many human disease such as inflammatory bowel disease (IBD) [13], chronic periodontitis [14], cancer [15, 16], psoriasis [17], several neurological disorders [18], et al. In relation to NONFH, elevated TNF-α serum concentration and expression of TNF-α in the bone marrow have been confirmed in both animal and clinical experiments during the process of steroid-induced osteonecrosis [19, 20]. The aggregation of macrophages into the necrotic area, resulting in the release of TNF-α together with other inflammatory factors, reinforces the inflammatory response and aggravates local osteoclasts [21].
TNF-α gene is located in the class III region of the major histocompatibility complex (MHC) and maps to 6p21.3 [22, 23]. It is revealed that gene polymorphisms in the regulatory region can generate the expression of TNF-α [22]. Recently, many genetic polymorphisms have been detected in the cytokine promoter at the initial position of TNF-α transcription site, which are 1031(T/C), 376(G/A), 308(G/A), 238(G/A), et al. [24]. Various scientific literatures have demonstrated TNF-α gene polymorphisms being associated to the susceptibility of systemic lupus erythematosus (SLE) [25], ankylosing spondylitis (AS) [26], gastric and hepatocellular carcinomas [27], and periodontitis [28], as well as NONFH [3, 29,30,31,32,33]. Though many relevant research programs have been performed, when it comes to the relation of TNF-α-308(G/A) (rs1800629) and -238(G/A) (rs361525) polymorphisms to NONFH susceptibility, no common consensus has been reached yet and there does not exist any meta-analysis summarizing those relative studies. Therefore, we decided to perform a meta-analysis focusing on the association between TNF-α-308(G/A) (rs1800629) and -238(G/A) (rs361525) polymorphisms and susceptibility of NONFH to draw a precise evaluation of the relationship, which might be a potential treatment approach and could assist the early diagnosis of NONFH.
Materials and methods
Identification of eligible studies
Relevant available literatures published in the PubMed, Web of Science, Embase, Cochrane library databases, China National Knowledge Infrastructure (CNKI), WANFANG Data, and China Science and Technology Journal Database (CSTJ) were searched up to January 30, 2018, by two investigators independently. We used the following retrieval terms: (“femoral head necrosis” or “FHN” or “osteonecrosis of the femoral head” or “ONFH” or “non-traumatic osteonecrosis of the femoral head” or “NONFH”) and (“TNF-α” or “tumour necrosis factor alpha” or “tumour necrosis factor α” or “TNF alpha” or “rs1800629” or “rs361525”) and (“polymorphi*” or “allele” or “genetic variant” or “gene*”). No language, race, ethnicity, or geographic area restrictions were applied. We also searched the bibliography of recent related reviews and the primary articles manually for all identified studies.
Inclusion and exclusion criteria
The inclusion criterion was formulated as follows: (1) the study was a case-control study design; (2) the study investigated the association between TNF-α gene-238 or -308 polymorphism and the risk of NONFH; (3) genotype frequencies of TNF-α gene-238 or -308 polymorphism were reported in the study; (4) the study was performed on human.
The studies were deemed inadequately if (1) they were case reports, reviews, descriptive studies, comments, or animal studies; (2) studies contained relevant data which had been published before; (3) studies did not provide available genotype frequencies and cannot be obtained by contacting with the authors; (4) there were obvious errors in research designs and statistical approaches.
All relevant literatures were appraised and discussed to reach a consensus by two investigators according to the inclusion and exclusion criteria independently.
Data extraction
Based on a standard form, two investigators extracted following information from all qualified studies independently: (1) name of the first author, (2) publication data, (3) population distribution (country and ethnicity), (4) gender and age of cases and controls, (5) numbers and diagnosis of case group and control group, (6) genotyping method, (7) genotype frequency in case group and control group. No remaining disagreement presents about the data among all authors.
Methodological quality assessment
Clark scores system, which contains ten items [34], was implemented to assess the qualities of the included studies by two investigators independently. Scores below 5 were regarded as low quality, while 5–7 scores indicate moderate quality and 8–10 scores represent high quality [34].
Statistical analysis
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement carefully in the whole process of this analysis [35]. By merging ORs with 95% CI, we evaluated the association between TNF-α polymorphisms and NONFH risks. Allelic frequencies of TNF-a gene-238 or -308 polymorphism from the respective studies were determined by the allele counting method. At first, the association strength between the allele and the variation to NONFH was examined (G versus A). Then, the pooled ORs and corresponding 95%CIs were computed in homozygote model (GG versus AA), heterozygote model (GA versus AA), dominant model (GG + GA versus AA), and recessive (GG versus GA + AA) model, respectively.
To determine statistical heterogeneity, Q statistic and I2 statistic were evaluated in each model. If P < 0.10 and I2 > 50%, significant heterogeneity was considered to exist [36]. The DerSimonian-Laird method for random effects model was used if significant heterogeneity existed, or the Mantel-Haenszel method for fixed effects model was chosen otherwise [37]. Due to the limitations of funnel plotting, which requires a range of studies with varying sizes and subjective judgments, we assessed publication bias using the Begg’s funnel plot and Egger’s weighted regression method as well as funnel plots. All analyses were conducted with STATA 14 (Stata, CollegeStation, TX). The P value (< 0.05) in Z test was considered to determine the significance of the pooled OR, and the result was two-sided.
Results
Characteristics of the included studies
We identified 329 records in total, retrieving from the database (PubMed = 26, WOS = 105, Embase = 28, Cochrane = 3, CNKI = 51, WANFANG Data = 57, CSTJ = 6) and reference lists (manual search = 53) (Fig. 1). After inspecting titles or abstracts and removing the duplicates and irrelevant articles, we obtained 23 studies for full-text examination. In terms of exclusion and inclusion criteria, 18 studies were eliminated (one was review, one did not present genotype frequencies, one’s full text was not available, eight were not about genetic polymorphisms, and seven contained no data of the target SNPs or diseases). The remaining five studies containing 432 NONFH patients and 760 controls were finally included in the analysis.
Characteristics of the five researches are displayed in Table 1. The aetiological diagnosis of osteonecrosis of the femoral head and the number of relative cases and controls was not clear in some of the eligible studies, but all the involved patients were corresponding to the diagnostic criteria of NONFH described within the studies, which has been inspected cautiously. In addition, it should be noted that 37 controls recruited in Shixin Wang et al. [33] were cured SARS patients with no femoral head necrosis, while the other 723 controls were all described as healthy individuals. Therefore, we used Hardy-Weinberg equilibrium (HWE) to analyze the genotype distribution and assess the reliability of subjects’ selection for each study. Although the selection of controls in S. Samara et al. [32] was likely to have poor quality (P < 0.05), the genotype distribution of the controls in Shixin Wang et al. [33] did not significantly vary from the natural populations (P > 0.05) (Table 2), which indicated that the selection of controls in in Shixin Wang et al. [33] was acceptable for our analysis.
Association between TNF-α-308 polymorphism and NONFH
Fixed pooling model was implemented in all the modes when we processed meta-analysis, and there were no significant heterogeneity revealed in any models. After pooling the genotype distribution of the 432 NONFH patients and 760 controls, we found out that significant associations were confirmed in overall population for alleles mode (G versus A) (OR = 0.726, 95% CI 0.566–0.933, P = 0.012, Fig. 2a) and recessive mode (GG versus GA/AA) (OR = 0.725, 95% CI 0.543–0.967, P = 0.029, Fig. 2d). As for homozygote mode (GG versus AA) (OR = 0.524, 95% CI 0.261–1.051, P = 0.069, Fig. 2b), heterozygote mode (GA versus AA) (OR = 0.682, 95% CI 0.319–1.459, P = 0.324), and dominant mode (GG/GA versus AA) (OR = 0.544, 95% CI 0.273–1.087, P = 0.085, Fig. 2c), there were no statistical significance identified in all the three modes overall (Table 3).
In the subgroup meta-analysis, evaluating strategy was stratified by population distribution (Asian and Caucasian). Fixed-effects mode was also applied since no significant heterogeneity was detected in any subgroups and analyzing modes. In Asian group, it was shown that TNF-α-308(G/A) polymorphism was significantly relative to susceptibility of NONFH in four modes consisting of alleles mode (OR = 0.648, 95% CI 0.475–0.885, P = 0.006,Fig. 2a), recessive mode (OR = 0.674, 95% CI 0.468–0.971, P = 0.034, Fig. 2d), homozygote mode (OR = 0.330, 95% CI 0.136–0.802, P = 0.014, Fig. 2b), and dominant mode (OR = 0.344, 95% CI 0.143–0.827, P = 0.017, Fig. 2c), but no significant association was observed in heterozygote mode (OR = 0.405, 95% CI 0.154–1.063, P = 0.066). On the contrary, in Caucasian group, the assessment of all five modes revealed no significance at all, which were alleles mode (OR = 0.909, 95% CI 0.590–1.400, P = 0.665), homozygote mode (OR = 1.842, 95% CI 0.386–8.793, P = 0.444), heterozygote mode (OR = 2.431, 95% CI 0.466–12.679, P = 0.292), recessive mode (OR = 0.823, 95% CI 0.511–1.323, P = 0.421), and dominant mode (OR = 1.966, 95% CI 0.408–9.473, P = 0.399) (Table 3).
Association between TNF-α-238 polymorphism and NONFH
As shown in Table 2, only three recruited studies provided the genotype and allelic distribution of TNF-α-238(G/A) polymorphism and there were 275 cases and 610 controls. In all the studies, neither NONFH patients nor controls were found carrying AA genotype of TNF-α-238 (Table 3), urging the utilization of alleles modes (G versus A) and recessive mode (GG versus GA) to demonstrate the connection. Similarly, homogeneity was achieved among the statistics and fixed-effects mode was chosen for the meta-analysis (Table 3). As a result, there was a significant association in alleles mode (OR = 0.270, 95% CI 0.4148–0.490, P < 0.001, Fig. 3a) as well as recessive mode (OR = 0.254, 95% CI 0.138–0.468, P < 0.001, Fig. 3b).
Sensitivity analysis
This meta-analysis adopted an exclusive strategy that was performed by ruling one research out each turn to draw the pooling ORs, 95% CIs, and Ps exclusively in each genetic mode.
For TNF-α-308(G/A) polymorphism, the ORs’ significance altered when we removed S. Samara et al. [32] in homozygote mode (GG versus GA, OR = 0.347, 95% CI 0.149–0.805, P = 0.014, Fig. 4b) and dominant mode (GG + GA versus AA, OR = 0.365, 95% CI 0.159–0.841, P = 0.018, Fig. 4c). We also observed that the ORs’ significance changed when we dropped Yaosheng Liu et al. [30] in alleles mode (G versus A, OR = 0.832, 95% CI 0.618–1.121, P = 0.228, Fig. 4a) and recessive mode (GG versus GA + AA, OR = 0.767, 95% CI 0.549–1.071, P = 0.119, Fig. 4d). We also spotted the results of recessive mode altered when deleting Biao-Fang Wei et al. [29] (OR = 0.742, 95% CI 0.520–1.060, P = 0.101, Fig. 4d) and Sanja Srzentić et al. [31] (OR = 0.751, 95% CI 0.555–1.018, P = 0.065, Fig. 4d).
For TNF-α-238(G/A) polymorphism, the ORs in any genetic mode analyzing did not change, indicating the stability of the results.
Publication bias
Funnel plots, Begg’s rank correlation method and Egger’s weighted regression method were applied to determine the publication bias. Though their power is relatively inadequate (especially with insufficient researches) [38, 39], we tried those approaches because of no better alternative methods. After analyzing all the genotype modes, it was reported that funnel plots were symmetric and the possible intersection of regression lines and y-axis included the origin. Furthermore, Begg’s and Egger’s tests suggested no significant publication bias (P > 0.10).
Discussion
The risk factors for NONFH include internal and external elements, such as age, hypoxia, corticosteroid use, alcohol intake, smoking, and various chronic diseases (renal disease, haematological disease, inflammatory bowel disease, post-organ transplantation, and hypertension) [32, 40, 41]. Many factors were involved in the aetiology of NONFH, among which evidences supporting the TNF-α gene-308 or -238 polymorphisms have been provided currently [29,30,31,32,33]. However, these distinct studies presented conclusions inconsistently. Therefore, we carried out this meta-analysis to reveal the association between TNF-α gene-308 or -238 polymorphisms and NONFH susceptibility.
Previous studies revealed that the TNF-α gene-308 polymorphism could be related to TNF-α gene regulation and associated with increased transcriptional activity of its product [32, 42, 43]. In our meta-analysis, overall samples revealed significant associations between TNF-α gene-308(G/A) polymorphism and NONFH susceptibility in alleles mode (G versus A) and recessive mode (GG versus GA/AA) (Table 3 and Fig. 2), which indicated that G allele may be a protective factor in the risk of NONFH. However, by comparing genotype GG with GA + AA, Biao-Fang Wei et al. found that the TNF-α mRNA and the TNF-α cytokine expression were not significantly different [29]. Thus, in-depth studies were required to identify the effects of TNF-α gene-308(G/A) polymorphisms on the development of NONFH. After analyzing by ethnicity, we found significant association in Asians but not in Caucasians, which suggested that the role TNF-α gene-308 polymorphism played in the risk of NONFH in Asian population was more notable than that in Caucasian population. The reasons why the SNP polymorphism in the same position of TNF-α gene played absolutely different roles in different races may lie on two aspects. First, the genetic backgrounds may differ from the two ethnic groups. Besides, there are numerous aetiological factors contributing to NONFH such as hyperlipidemia, autoimmune diseases, alcoholism, et al. [7, 8]. The varied prevalence of hyperlipidemia [44], autoimmune diseases [45], discrepant alcohol consumption [46, 47], et al., in different populations may explain the inconsistency. By the way, due to the fact that P for HWE was too small (Table 2) in study conducted by S. Samara et al. [32], we removed it in the sensitivity analysis. Consequently, homozygote mode (OR = 0.347, 95% CI 0.149–0.805, P = 0.014, Fig. 4b) and dominant mode (OR = 0.365, 95% CI 0.159–0.841, P = 0.018, Fig. 4c) showed different results, which indicated that the improper selection of the controls labilized the pooled estimate. Nevertheless, the deletion of Samara et al. [32] increased the number of mode type that revealed significant association.
Another common SNP of TNF-α we paid attention to is TNF-α gene-238G/A polymorphism. Samara et al. reported a G-to-A change at position 238 resulted in an up-regulation of TNF-α gene expression and suggested that the increase of TNF-α expression could lead to osteoclasts proliferation, which contributed to NONFH [32]. On the contrary, another study recently showed that the A-238 allele at position 238 may be associated with downregulation of tissue inflammation [43]. We conducted a meta-analysis including three eligible case-control studies containing 275 cases comparing 610 controls. The allele A was so rare that neither NONFH patients nor controls were found carrying AA genotype of TNF-α gene-238. For this reason, only the alleles mode (G vs. A) and recessive mode (GG vs. (GA + AA)) were conducted in the meta-analysis. The total sample demonstrated that statistical significance existed in both of the two models. The allele A at position 238 could be a risk factor to NONFH.
Tumour necrosis factor (TNF-α) gene is located on human chromosome 6p21.3 in the vicinity of the major histocompatibility complex (MHC) class III region [48] and plays an important role in inflammation, immunity, and cellular organization of its product. As we discussed above, the TNF-α gene-308 or -238 affected regulation of TNF-α gene and associated with altered transcriptional activity in many diseases. What is more, a number of studies had shown that the TNF-α promoter polymorphisms have a vital effect on transcriptional activity [49, 50]. It is generally believed that the pathophysiology of NONFH is related to the apoptosis of osteoblasts and osteocytes. Shibahara et al. suggested that there were mass apoptosis cells in necrotic zone and the apoptosis of osteocytes resulted in the osteonecrosis and the destruction of bone structure [51]. Besides, it is known that some cytokines such as IL, TNF-α, TNF-β, and other inflammatory-related cytokines play an important role in the balance between osteoclasts ad osteoblasts. Dai CY et al. have reported that TNF-α acts on osteoblasts or bone marrow cells to synthesize and release cytokines which is directly associated with osteoclasts proliferation and maturation [52]. Thus, we could assume that the TNF-α gene-308 or -238 polymorphisms led to an altered expression of the gene, which made a contribution to inflammation response. Consequently, apoptosis of osteoblasts and proliferation of osteoclasts were activated or aggravated, resulting in the deterioration of NONFH.
To the best of our knowledge, this is the first meta-analysis to comprehensively assess the association between TNF-α gene-308(G/A) and -238(G/A) and the susceptibility of NONFH, which may provide the clues of prevention and early diagnosis and potential trials for osteonecrosis treatment. During the searching process, we have followed specific and repeatable strategies and applied strict inclusion and exclusion criteria to recruit. We also considered the distribution of ethnic groups and applied subgroup analysis to assess the association in Asian and Caucasian populations, respectively. Furthermore, Clark scores was used to assess the quality of each studies and most of them possessed high qualities. HWE was used to test the homogeneity between controls and natural populations, and only one study was significantly deviated. Besides, no significant heterogeneity was identified among the enrolled studies. Despite of the insufficient research number, we still applied the Egger’s and Begg’s test and found no publication bias in the recruitment.
Nevertheless, a few limitations shall be claimed in this meta-analysis. First, the language was limited to English and Chinese, which may develop bias as a consequence of the restrictedly defined populations. Second, only two out of five studies mentioned the severity of NONFH. Thus, the limited number defies further analysis of the associations between TNF-α gene-308(G/A) and -238(G/A) polymorphisms and clinical stages or features of NONFH. Third, following the enrollment strategies, we excluded some researches due to lack of target SNPs data, which may increase the potential of selection bias. Fourth, in the sensitivity analysis, the ORs’ significance changed for TNF-α gene-308(G/A) polymorphism in homozygote mode and dominant mode after we removed S. Samara et al. [32]. As shown in Table 2, the genotyping variance of control group in S. Samara et al. [32] was derived from HWE, which might be the reason of the unstable results. Additionally, the removal of Yaosheng Liu et al. [30] also influenced the ORs’ significance for TNF-α gene-308(G/A) polymorphism in alleles mode and recessive mode. The weights of Yaosheng Liu et al. [30] in the meta-analysis of alleles mode and recessive mode were much heavy, which were 34.46 and 27.93%, respectively. Thus, the significant effects of this research indicate that further original studies are necessary to enhance the result stability.
Conclusion
This meta-analysis shows that TNF-α-308(G/A) and -238(G/A) polymorphisms are inversely associated with the risk of NONFH, while the significant association for 308(G/A) is observed in Asian. Original and intensive researches of individuals of diverse ethnicities would be essential due to the insufficient or conflict interpretations of the polymorphism effects on the NONFH pathology.
Reference
Agrawal K, Tripathy SK, Sen RK et al (2017) Nuclear medicine imaging in osteonecrosis of hip: old and current concepts. World J Orthop 8(10):747–753. https://doi.org/10.5312/wjo.v8.i10.747
Filipowska J, Tomaszewski KA, Niedzwiedzki L et al (2017) The role of vasculature in bone development, regeneration and proper systemic functioning. Angiogenesis 20(3):291–302. https://doi.org/10.1007/s10456-017-9541-1
Wang C, Wang Y, Meng HY et al (2015) Application of bone marrow mesenchymal stem cells to the treatment of osteonecrosis of the femoral head. Int J Clin Exp Med 8(3):3127–3135
Saidi S, Magne D (2011) Interleukin-33: a novel player in osteonecrosis of the femoral head? Joint Bone Spine 78(6):550–554. https://doi.org/10.1016/j.jbspin.2011.04.013
Zhang Y, Sun R, Zhang L et al (2017) Effect of blood biochemical factors on nontraumatic necrosis of the femoral head: logistic regression analysis. Orthopade 46(9):737–743. https://doi.org/10.1007/s00132-017-3408-4
Celik A, Tekis D, Saglam F et al (2006) Association of corticosteroids and factor V, prothrombin, and MTHFR gene mutations with avascular osteonecrosis in renal allograft recipients. Transplant Proc 38(2):512–516. https://doi.org/10.1016/j.transproceed.2005.12.062
Malizos KN, Karantanas AH, Varitimidis SE et al (2007) Osteonecrosis of the femoral head: etiology, imaging and treatment. Eur J Radiol 63(1):16–28. https://doi.org/10.1016/j.ejrad.2007.03.019
Etienne G, Mont MA, Ragland PS (2004) The diagnosis and treatment of nontraumatic osteonecrosis of the femoral head. Instr Course Lect 53:67–85
Mont MA, Cherian JJ, Sierra RJ et al (2015) Nontraumatic osteonecrosis of the femoral head: where do we stand today? A ten-year update. J Bone Joint Surg Am 97(19):1604–1627. https://doi.org/10.2106/JBJS.O.00071
Carswell EA, Old LJ, Kassel RL et al (1975) An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A 72(9):3666–3670
Olszewski MB, Groot AJ, Dastych J et al (2007) TNF trafficking to human mast cell granules: mature chain-dependent endocytosis. J Immunol 178(9):5701–5709
Hayashi K, Piras V, Tabata S et al (2013) A systems biology approach to suppress TNF-induced proinflammatory gene expressions. Cell Commun Signal 11:84. https://doi.org/10.1186/1478-811X-11-84
Argollo M, Fiorino G, Hindryckx P et al (2017) Novel therapeutic targets for inflammatory bowel disease. J Autoimmun 85:103–116. https://doi.org/10.1016/j.jaut.2017.07.004
Cardoso EM, Reis C, Manzanares-Cespedes MC (2018) Chronic periodontitis, inflammatory cytokines, and interrelationship with other chronic diseases. Postgrad Med 130(1):98–104. https://doi.org/10.1080/00325481.2018.1396876
Ahmad S, Khan M Y, Rafi Z, et al(2017) Oxidation, glycation and glycoxidation—the vicious cycle and lung cancer. Semin Cancer Biol. https://doi.org/10.1016/j.semcancer.2017.10.005
Pandey MK, Gupta SC, Nabavizadeh A et al (2017) Regulation of cell signaling pathways by dietary agents for cancer prevention and treatment. Semin Cancer Biol 46:158–181. https://doi.org/10.1016/j.semcancer.2017.07.002
Kim TG, Kim SH, Lee MG (2017) The origin of skin dendritic cell network and its role in psoriasis. Int J Mol Sci 19(1). https://doi.org/10.3390/ijms19010042
Olmos G, Llado J (2014) Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediat Inflamm 2014:861231. https://doi.org/10.1155/2014/861231
Scheper MA, Badros A, Chaisuparat R et al (2009) Effect of zoledronic acid on oral fibroblasts and epithelial cells: a potential mechanism of bisphosphonate-associated osteonecrosis. Br J Haematol 144(5):667–676. https://doi.org/10.1111/j.1365-2141.2008.07504.x
Okazaki S, Nishitani Y, Nagoya S et al (2009) Femoral head osteonecrosis can be caused by disruption of the systemic immune response via the toll-like receptor 4 signalling pathway. Rheumatology (Oxford) 48(3):227–232. https://doi.org/10.1093/rheumatology/ken462
Wu X, Feng X, He Y et al (2016) IL-4 administration exerts preventive effects via suppression of underlying inflammation and TNF-alpha-induced apoptosis in steroid-induced osteonecrosis. Osteoporos Int 27(5):1827–1837. https://doi.org/10.1007/s00198-015-3474-6
Zhang BB, Liu XZ, Sun J et al (2013) Association between TNF alpha gene polymorphisms and the risk of duodenal ulcer: a meta-analysis. PLoS One 8(2):e57167. https://doi.org/10.1371/journal.pone.0057167
Nedwin GE, Naylor SL, Sakaguchi AY et al (1985) Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic Acids Res 13(17):6361–6373
Elahi MM, Asotra K, Matata BM et al (2009) Tumor necrosis factor alpha-308 gene locus promoter polymorphism: an analysis of association with health and disease. Biochim Biophys Acta 1792(3):163–172
Piotrowski P, Wudarski M, Sowinska A et al (2015) TNF-308 G/A polymorphism and risk of systemic lupus erythematosus in the polish population. Mod Rheumatol 25(5):719–723. https://doi.org/10.3109/14397595.2015.1008778
Manolova I, Ivanova M, Stoilov R et al (2014) Association of single nucleotide polymorphism at position -308 of the tumor necrosis factor-alpha gene with ankylosing spondylitis and rheumatoid arthritis. Biotechnol Biotechnol Equip 28(6):1108–1114. https://doi.org/10.1080/13102818.2014.972147
Guo XF, Wang J, Yu SJ et al (2013) TNF-alpha-308 polymorphism and risk of digestive system cancers: a meta-analysis. World J Gastroenterol 19(48):9461–9471. https://doi.org/10.3748/wjg.v19.i48.9461
Xie CJ, Chen L, Tong FL et al (2012) Meta analysis of association between TNF-alpha-308 polymorphism and periodontitis in Chinese Han population. Shanghai Kou Qiang Yi Xue 21(4):447–450
Wei BF, Feng Z, Wei W et al (2017) Associations of TNF-alpha −238 a/G and IL-10-1082 G/a genetic polymorphisms with the risk of NONFH in the Chinese population. J Cell Biochem 118(12):4872–4880. https://doi.org/10.1002/jcb.26167
Liu Y, Jiang W, Liu S et al (2015) Combined effect of tnf-alpha polymorphisms and hypoxia on steroid-induced osteonecrosis of femoral head. Int J Clin Exp Pathol 8(3):3215–3219
Srzentic S, Nikcevic G, Spasovski D et al (2015) Predictive genetic markers of coagulation, inflammation and apoptosis in Perthes disease-Serbian experience. Eur J Pediatr 174(8):1085–1092. https://doi.org/10.1007/s00431-015-2510-z
Samara S, Kollia P, Dailiana Z et al (2012) Predictive role of cytokine gene polymorphisms for the development of femoral head osteonecrosis. Dis Markers 33(4):215–221. https://doi.org/10.3233/DMA-2012-0928
Wang S, Wei M, Han Y et al (2008) Roles of TNF-alpha gene polymorphisms in the occurrence and progress of SARS-Cov infection: a case-control study. BMC Infect Dis 8:27. https://doi.org/10.1186/1471-2334-8-27
Srivastava K, Srivastava A, Sharma KL et al (2011) Candidate gene studies in gallbladder cancer: a systematic review and meta-analysis. Mutat Res 728(1–2):67–79. https://doi.org/10.1016/j.mrrev.2011.06.002
Vrabel M (2015) Preferred reporting items for systematic reviews and meta-analyses. Oncol Nurs Forum 42(5):552–554. https://doi.org/10.1188/15.ONF.552-554
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558. https://doi.org/10.1002/sim.1186
DerSimonian R, Laird N (2015) Meta-analysis in clinical trials revisited. Contemp Clin Trials 45(Pt A):139–145. https://doi.org/10.1016/j.cct.201509.002
Papageorgiou SN, Papadopoulos MA, Athanasiou AE (2014) Assessing small study effects and publication bias in orthodontic meta-analyses: a meta-epidemiological study. Clin Oral Investig 18(4):1031–1044. https://doi.org/10.1007/s00784-014-1196-3
Ioannidis JP, Trikalinos TA (2007) The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. CMAJ 176(8):1091–1096. https://doi.org/10.1503/cmaj.060410
Guo KJ, Zhao FC, Guo Y et al (2014) The influence of age, gender and treatment with steroids on the incidence of osteonecrosis of the femoral head during the management of severe acute respiratory syndrome: a retrospective study. Bone Joint J 96-B(2):259–262. https://doi.org/10.1302/0301-620X.96B2.31935
Zou W, Yang S, Zhang T et al (2015) Hypoxia enhances glucocorticoid-induced apoptosis and cell cycle arrest via the PI3K/Akt signaling pathway in osteoblastic cells. J Bone Miner Metab 33(6):615–624. https://doi.org/10.1007/s00774-014-0627-1
Jiang Y, Wang X, Cheng Y et al (2017) Associations between inflammatory gene polymorphisms (TNF-alpha 308G/A, TNF-alpha 238G/A, TNF-beta 252A/G, TGF-beta1 29T/C, IL-6 174G/C and IL-10 1082A/G) and susceptibility to osteosarcoma: a meta-analysis and literature review. Oncotarget 8(57):97571–97583. https://doi.org/10.18632/oncotarget.18813
Sghaier I, Zidi S, Mouelhi L et al (2015) The relationship between TNF alpha gene polymorphisms (-238/-308), TNF RII VNTR (p75) and outcomes of hepatitis B virus infection in Tunisian population. Gene 568(2):140–145. https://doi.org/10.1016/j.gene.2015.05.029
Younossi ZM, Koenig AB, Abdelatif D et al (2016) Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64(1):73–84. https://doi.org/10.1002/hep.28431
Tiffin N, Hodkinson B, Okpechi I (2014) Lupus in Africa: can we dispel the myths and face the challenges? Lupus 23(1):102–111. https://doi.org/10.1177/0961203313509296
Guo L, Deng J, He Y et al (2016) Alcohol use and alcohol-related problems among adolescents in China: a large-scale cross-sectional study. Medicine (Baltimore) 95(38):e4533. https://doi.org/10.1097/MD.0000000000004533
Moure-Rodriguez L, Caamano-Isorna F, Doallo S et al (2014) Heavy drinking and alcohol-related injuries in college students. Gac Sanit 28(5):376–380. https://doi.org/10.1016/j.gaceta.2014.02.017
Hajeer AH, Hutchinson IV (2000) TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech 50(3):216–228. https://doi.org/10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.0.CO;2-Q
Verma S, Slutsky AS (2007) Idiopathic pulmonary fibrosis—new insights. New Engl J Med 356(13):1370–1372. https://doi.org/10.1056/NEJMcibr070490
Kroeger KM, Carville KS, Abraham LJ (1997) The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol 34(5):391–399
Shibahara M, Nishida K, Asahara H et al (2000) Increased osteocyte apoptosis during the development of femoral head osteonecrosis in spontaneously hypertensive rats. Acta Med Okayama 54(2):67–74. https://doi.org/10.18926/AMO/32287
Dai CY, Chuang WL, Lee LP et al (2006) Associations of tumour necrosis factor alpha promoter polymorphisms at position -308 and -238 with clinical characteristics of chronic hepatitis C. J Viral Hepat 13(11):770–774. https://doi.org/10.1111/j.1365-2893.2006.00767.x
Acknowledgements
This study was supported by the National Key Research and Development Program of China (2016YFC1100100) and the Major Research Plan of National Natural Science Foundation of China (No. 91649204).
Funding
This study was funded by the National Key Research and Development Program of China (2016YFC1100100) and the Major Research Plan of National Natural Science Foundation of China (No. 91649204).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Informed consent
All five recruited studies declared that the informed consent was obtained from all individuals included in the studies.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Peng, Y., Liu, Y., Huang, D. et al. Association of TNF-α-308(G/A) and -238(G/A) polymorphisms with non-traumatic osteonecrosis of the femoral head risks: a meta-analysis. International Orthopaedics (SICOT) 42, 1711–1721 (2018). https://doi.org/10.1007/s00264-018-3859-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00264-018-3859-1