Abstract
Background
Analysis of hepatocellular carcinoma (HCC) single-cell sequencing data was conducted to explore the role of tumor-associated neutrophils in the tumor microenvironment.
Methods
Analysis of single-cell sequencing data from 12 HCC tumor cores and five HCC paracancerous tissues identified cellular subpopulations and cellular marker genes. The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases were used to establish and validate prognostic models. xCELL, TIMER, QUANTISEQ, CIBERSORT, and CIBERSORT-abs analyses were performed to explore immune cell infiltration. Finally, the pattern of tumor-associated neutrophil roles in tumor microenvironmental components was explored.
Results
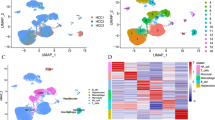
A total of 271 marker genes for tumor-associated neutrophils were identified based on single-cell sequencing data. Prognostic models incorporating eight genes were established based on TCGA data. Immune cell infiltration differed between the high- and low-risk groups. The low-risk group benefited more from immunotherapy. Single-cell analysis indicated that tumor-associated neutrophils were able to influence macrophage, NK cell, and T-cell functions through the IL16, IFN-II, and SPP1 signaling pathways.
Conclusion
Tumor-associated neutrophils regulate immune functions by influencing macrophages and NK cells. Models incorporating tumor-associated neutrophil-related genes can be used to predict patient prognosis and immunotherapy responses.








Similar content being viewed by others
Data availability
Publicly available datasets were analyzed in this study.
Abbreviations
- GEO:
-
Gene expression omnibus
- GSEA:
-
Gene set enrichment analysis
- HCC:
-
Hepatocellular carcinoma
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LASSO:
-
Least absolute shrinkage and selection operator
- RT-qPCR:
-
Real-time quantitative PCR
- ROC:
-
Receiver operating characteristic
- ssGSEA:
-
Single-sample GSEA
- TCGA:
-
The cancer genome atlas program
- UMAP:
-
Uniform manifold approximation and projection
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Huang DQ, Singal AG, Kono Y, Tan DJH, El-Serag HB, Loomba R (2022) Changing global epidemiology of liver cancer from 2010 to 2019: NASH is the fastest growing cause of liver cancer. Cell Metab 34(7):969–77.e2
Liu Z, Jiang Y, Yuan H, Fang Q, Cai N, Suo C et al (2019) The trends in incidence of primary liver cancer caused by specific etiologies: results from the global burden of disease study 2016 and implications for liver cancer prevention. J Hepatol 70(4):674–683
Liu CY, Chen KF, Chen PJ (2015) Treatment of liver cancer. Cold Spring Harb Perspect Med 5(9):a021535
Grady WM, Yu M, Markowitz SD (2021) Epigenetic alterations in the gastrointestinal tract: current and emerging use for biomarkers of cancer. Gastroenterology 160(3):690–709
Chowdhury MMH, Salazar CJJ, Nurunnabi M (2021) Recent advances in bionanomaterials for liver cancer diagnosis and treatment. Biomaterials science 9(14):4821–4842
Zhao C, Liu S, Gao F, Zou Y, Ren Z, Yu Z (2022) The role of tumor microenvironment reprogramming in primary liver cancer chemotherapy resistance. Front Oncol 12:1008902
Polidoro MA, Mikulak J, Cazzetta V, Lleo A, Mavilio D, Torzilli G et al (2020) Tumor microenvironment in primary liver tumors: a challenging role of natural killer cells. World J Gastroenterol 26(33):4900–4918
Zhang QY, Ho DW, Tsui YM, Ng IO (2022) Single-cell transcriptomics of liver cancer: hype or insights? Cell Mol Gastroenterol Hepatol 14(3):513–525
Kotsiliti E, Leone V, Schuehle S, Govaere O, Li H, Wolf MJ et al (2023) Intestinal B cells license metabolic T-cell activation in NASH microbiota/antigen-independently and contribute to fibrosis by IgA-FcR signalling. J Hepatol 79(2):296–313
Qin R, Zhao H, He Q, Li F, Li Y, Zhao H (2022) Advances in single-cell sequencing technology in the field of hepatocellular carcinoma. Front Genet 13:996890
Ma L, Heinrich S, Wang L, Keggenhoff FL, Khatib S, Forgues M et al (2022) Multiregional single-cell dissection of tumor and immune cells reveals stable lock-and-key features in liver cancer. Nat Commun 13(1):7533
Zhao N, Dang H, Ma L, Martin SP, Forgues M, Ylaya K et al (2021) Intratumoral γδ T-cell infiltrates, chemokine (C-C Motif) ligand 4/chemokine (C-C Motif) ligand 5 protein expression and survival in patients with hepatocellular carcinoma. Hepatology (Baltimore, MD) 73(3):1045–1060
Scheiner B, Pinato DJ (2023) Tumor-infiltrating neutrophils: gatekeepers in liver cancer immune control. Gastroenterology 164(7):1338–1339
Wu F, Fan J, He Y, Xiong A, Yu J, Li Y et al (2021) Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun 12(1):2540
Xue R, Zhang Q, Cao Q, Kong R, Xiang X, Liu H et al (2022) Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 612(7938):141–147
Forner A, Reig M, Bruix J (2018) Hepatocellular carcinoma. Lancet (London, England) 391(10127):1301–1314
Yang JD, Heimbach JK (2020) New advances in the diagnosis and management of hepatocellular carcinoma. BMJ (Clinical Research Ed) 371:m3544
Bruix J, Reig M, Sherman M (2016) Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology 150(4):835–853
Oura K, Morishita A, Tani J, Masaki T (2021) Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: a review. Int J Mol Sci 22(11):5801
Wang X, Hassan W, Jabeen Q, Khan GJ, Iqbal F (2018) Interdependent and independent multidimensional role of tumor microenvironment on hepatocellular carcinoma. Cytokine 103:150–159
Giese MA, Hind LE, Huttenlocher A (2019) Neutrophil plasticity in the tumor microenvironment. Blood 133(20):2159–2167
Ng MSF, Tan L, Wang Q, Mackay CR, Ng LG (2021) Neutrophils in cancer-unresolved questions. Sci China Life Sci 64(11):1829–1841
Zhang X, Zhang W, Yuan X, Fu M, Qian H, Xu W (2016) Neutrophils in cancer development and progression: roles, mechanisms, and implications (Review). Int J Oncol 49(3):857–867
Chen H, Zhou XH, Li JR, Zheng TH, Yao FB, Gao B et al (2021) Neutrophils: driving inflammation during the development of hepatocellular carcinoma. Cancer Lett 522:22–31
Geh D, Leslie J, Rumney R, Reeves HL, Bird TG, Mann DA (2022) Neutrophils as potential therapeutic targets in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 19(4):257–273
Chen C, Wang Z, Ding Y, Qin Y (2023) Tumor microenvironment-mediated immune evasion in hepatocellular carcinoma. Front Immunol 14:1133308
Li XY, Shen Y, Zhang L, Guo X, Wu J (2022) Understanding initiation and progression of hepatocellular carcinoma through single cell sequencing. Biochim Biophys Acta 1877(3):188720
Arvanitakis K, Mitroulis I, Germanidis G (2021) Tumor-associated neutrophils in hepatocellular carcinoma pathogenesis, prognosis, and therapy. Cancers 13(12):2899
Liu L, Zhang R, Deng J, Dai X, Zhu X, Fu Q et al (2022) Construction of TME and Identification of crosstalk between malignant cells and macrophages by SPP1 in hepatocellular carcinoma. Cancer Immunol Immunother : CII 71(1):121–136
Liu Y, Xun Z, Ma K, Liang S, Li X, Zhou S et al (2023) Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J Hepatol 78(4):770–782
Hu J, Zhang L, Xia H, Yan Y, Zhu X, Sun F et al (2023) Tumor microenvironment remodeling after neoadjuvant immunotherapy in non-small cell lung cancer revealed by single-cell RNA sequencing. Genome Medicine 15(1):14
Eun JW, Yoon JH, Ahn HR, Kim S, Kim YB, Lim SB et al (2023) Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and lenvatinib. Cancer Commun (London, England) 43(4):455–479
Hu J, Yang L, Peng X, Mao M, Liu X, Song J et al (2022) ALDH2 hampers immune escape in liver hepatocellular carcinoma through ROS/Nrf2-mediated autophagy. Inflammation 45(6):2309–2324
Zhang H, Fu L (2021) The role of ALDH2 in tumorigenesis and tumor progression: targeting ALDH2 as a potential cancer treatment. Acta pharmaceutica Sinica B 11(6):1400–1411
Su Y, Xue C, Gu X, Wang W, Sun Y, Zhang R et al (2023) Identification of a novel signature based on macrophage-related marker genes to predict prognosis and immunotherapeutic effects in hepatocellular carcinoma. Front Oncol 13:1176572
Tao Z, Huang J, Li J. (2023) Comprehensive intratumoral heterogeneity landscaping of liver hepatocellular carcinoma and discerning of APLP2 in cancer progression. Environ Toxicol
Marengo A, Rosso C, Bugianesi E (2016) Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med 67:103–117
Tang J, Yan Z, Feng Q, Yu L, Wang H (2021) The roles of neutrophils in the pathogenesis of liver diseases. Front Immunol 12:625472
Wang X, Huang H, Sze KM, Wang J, Tian L, Lu J et al (2023) S100A10 promotes HCC development and progression via transfer in extracellular vesicles and regulating their protein cargos. Gut 72(7):1370–1384
Zhou X, Shi M, Cao J, Yuan T, Yu G, Chen Y et al (2021) S100 calcium binding protein A10, A novel oncogene, promotes the proliferation, invasion, and migration of hepatocellular carcinoma. Front Genet 12:695036
Mukai Y, Yamaguchi A, Sakuma T, Nadai T, Furugen A, Narumi K et al (2022) Involvement of SLC16A1/MCT1 and SLC16A3/MCT4 in l-lactate transport in the hepatocellular carcinoma cell line. Biopharm Drug Dispos 43(5):183–191
Jiang Z, Zhou X, Li R, Michal JJ, Zhang S, Dodson MV et al (2015) Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cellular Mol Life Sci : CMLS 72(18):3425–3439
Yang LY, Luo Q, Lu L, Zhu WW, Sun HT, Wei R et al (2020) Increased neutrophil extracellular traps promote metastasis potential of hepatocellular carcinoma via provoking tumorous inflammatory response. J Hematol Oncol 13(1):3
Du D, Liu C, Qin M, Zhang X, Xi T, Yuan S et al (2022) Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharmaceutica Sinica B 12(2):558–580
Sin SQ, Mohan CD, Goh RMW, You M, Nayak SC, Chen L, et al. (2022) Hypoxia signaling in hepatocellular carcinoma: challenges and therapeutic opportunities. Cancer metastasis reviews
Wang L, Liu Y, Dai Y, Tang X, Yin T, Wang C et al (2023) Single-cell RNA-seq analysis reveals BHLHE40-driven pro-tumour neutrophils with hyperactivated glycolysis in pancreatic tumour microenvironment. Gut 72(5):958–971
Xia S, Pan Y, Liang Y, Xu J, Cai X (2020) The microenvironmental and metabolic aspects of sorafenib resistance in hepatocellular carcinoma. EBioMedicine 51:102610
Zeng Q, Klein C, Caruso S, Maille P, Laleh NG, Sommacale D et al (2022) Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J Hepatol 77(1):116–127
Acknowledgements
None
Funding
This study was funded by the Hubei Provincial Fund Committee (2022CFB122).
Author information
Authors and Affiliations
Contributions
DY contributed to the conceptualization, JS, BY, and YS were involved in data curation, Yu Zhou and Yanbing Zhang contributed to the formal analysis, DY and Yu Zhou were involved in writing—original draft, and YD and KZ contributed to writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
None.
Ethical approval
This study was approved by the Ethics Committee of Renmin Hospital of Wuhan University. Written informed consent was obtained from all patients to participate in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, D., Zhou, Y., Zhang, Y. et al. Comprehensive analysis of scRNA-Seq and bulk RNA-Seq data reveals dynamic changes in tumor-associated neutrophils in the tumor microenvironment of hepatocellular carcinoma and leads to the establishment of a neutrophil-related prognostic model. Cancer Immunol Immunother 72, 4323–4335 (2023). https://doi.org/10.1007/s00262-023-03567-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-023-03567-4