Abstract
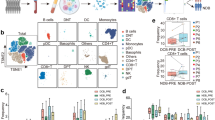
High expression of PD-L1 predicts PD-1/PD-L1 inhibitor benefit, meanwhile a few PD-L1-negative patients still benefit from these drugs. In this study, we aimed to explore the underlying cellular and molecular characteristics via single-cell sequencing. Before and after treatment with Pembrolizumab, peripheral blood mononuclear cells (PBMCs) were isolated via Ficoll gradient. Thereafter, single-cell RNA sequencing was performed, and clinical significance was validated with The Cancer Genome Atlas (TCGA) cohort. All 3423 cells of 16 clusters were classified into eight cell types, including NKG7+ T, NKG7+ NK, Naïve T, CDC1C+ dendritic cells, CD8+ T cells, B cells, macrophages and erythrocytes. Cell proportion, the clinical significance of differentially expressed genes and significant pathways of NKG7+ T, NKG7+ NK, Naïve T and CD8+ T cells were analyzed. Ubiquitin-mediated proteolysis/cell cycle/natural killer cell-mediated cytotoxicity were identified as PD-1 blockage-responsive pathways in NKG7+ NK cells. Apoptosis/Th1 and Th2 cell differentiation were proposed as Pembrolizumab-affected pathways in NKT cells. In gene level, ID2, PIK3CD, UQCR10, MATK, MZB1, IL7R and TRGC2 showed a significant correlation with PD-1 expression after TCGA dataset validation, which could possess potential as predictive markers for patients with PD-L1-negative lung squamous cell carcinoma who can benefit from Pembrolizumab.







Similar content being viewed by others
Availability of data and materials
All relevant data can be acquired by contacting the correspondent author.
Abbreviations
- FDA:
-
Food and Drug Administration
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- NK:
-
Natural killer
- NSCLC:
-
Non-small cell lung cancer
- PBMCs:
-
Peripheral blood mononuclear cells
- PD-1:
-
Programmed cell death 1
- PD-L1:
-
Programmed cell death 1 ligand 1
- TCGA:
-
The Cancer Genome Atlas
- t-SNE:
-
t-distributed Stochastic Neighbor Embedding
References
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics. CA Cancer J Clin 69(2019):7–34
Socinski MA, Obasaju C, Gandara D, Hirsch FR, Bonomi P, Bunn PA Jr, Kim ES, Langer CJ, Natale RB, Novello S, Paz-Ares L, Perol M, Reck M, Ramalingam SS, Reynolds CH, Spigel DR, Wakelee H, Thatcher N (2018) Current and emergent therapy options for advanced squamous cell lung cancer. J Thorac Oncol 13:165–183
Sunshine J, Taube JM (2015) PD-1/PD-L1 inhibitors. Curr Opin Pharmacol 23:32–38
Gettinger S, Rizvi NA, Chow LQ, Borghaei H, Brahmer J, Ready N, Gerber DE, Shepherd FA, Antonia S, Goldman JW, Juergens RA, Laurie SA, Nathan FE, Shen Y, Harbison CT, Hellmann MD (2016) Nivolumab monotherapy for the first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol 34:2980–2987
Ninomiya K, Hotta K (2018) Pembrolizumab for the first-line treatment of non-small cell lung cancer. Expert Opin Biol Ther 18:1015–1021
Chretien S, Zerdes I, Bergh J, Matikas A, Foukakis T (2019) Beyond PD-1/PD-L1 inhibition: what the future holds for breast cancer immunotherapy. Cancers (Basel) 11
Salmaninejad A, Valilou SF, Shabgah AG, Aslani S, Alimardani M, Pasdar A, Sahebkar A (2019) PD-1/PD-L1 pathway: basic biology and role in cancer immunotherapy. J Cell Physiol 234:16824–16837
Sun C, Mezzadra R, Schumacher TN (2018) Regulation and function of the PD-L1 checkpoint. Immunity 48:434–452
Sacher AG, Gandhi L (2016) Biomarkers for the clinical use of PD-1/PD-L1 inhibitors in non-small-cell lung cancer: a review. JAMA Oncol 2:1217–1222
Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X, Wu K (2018) Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer 17:129
Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR (2016) Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 375:1823–1833
Shen X, Zhao B (2018) Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: meta-analysis. BMJ 362:k3529
Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, Cortinovis DL, Leach J, Polikoff J, Barrios C, Kabbinavar F, Frontera OA, De Marinis F, Turna H, Lee JS, Ballinger M, Kowanetz M, He P, Chen DS, Sandler A, Gandara DR (2017) Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 389:255–265
Wang J, Chmielowski B, Pellissier J, Xu R, Stevinson K, Liu FX (2017) Cost-effectiveness of pembrolizumab versus ipilimumab in ipilimumab-naive patients with advanced melanoma in the United States. J Manag Care Spec Pharm 23:184–194
Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M, Choi K, Fromme RM, Dao P, McKenney PT, Wasti RC, Kadaveru K, Mazutis L, Rudensky AY, Pe’er D (2018) Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174:1293-1308.e1236
Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, Lauffenburger DA, Raue A (2018) Analysis of single-cell rna-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep 25:1458-1468.e1454
Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA (2016) Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352:189–196
Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, Yu EA, Schenk EL, Tan W, Zee A, Tan M, Gui P, Lea T, Wu W, Urisman A, Jones K, Sit R, Kolli PK, Seeley E, Gesthalter Y, Le DD, Yamauchi KA, Naeger DM, Bandyopadhyay S, Shah K, Cech L, Thomas NJ, Gupta A, Gonzalez M, Do H, Tan L, Bacaltos B, Gomez-Sjoberg R, Gubens M, Jahan T, Kratz JR, Jablons D, Neff N, Doebele RC, Weissman J, Blakely CM, Darmanis S, Bivona TG (2020) Therapy-Induced Evolution of Human Lung Cancer Revealed by Single-Cell RNA Sequencing. Cell 182:1232-12511e222
Fuss IJ, Kanof ME, Smith PD, Zola H (2009) Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. Chapter 7 Unit7 1
McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R (2013) Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29:461–467
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single-cell data. Cell 177(1888–1902):e1821
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16:284–287
Tang Z, Li C, Kang B, Gao G, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102
Dong W, Wu X, Ma S, Wang Y, Nalin AP, Zhu Z, Zhang J, Benson DM, He K, Caligiuri MA, Yu J (2019) The mechanism of Anti-PD-L1 antibody efficacy against PD-L1-negative tumors identifies NK cells expressing PD-L1 as a cytolytic effector. Cancer Discov 9:1422–1437
M. Lin, H. Luo, S. Liang, J. Chen, A. Liu, L. Niu, Y. Jiang, Pembrolizumab plus allogeneic NK cells in advanced non-small cell lung cancer patients. J Clin Invest (2020).
Kim H, Kwon HJ, Han YB, Park SY, Kim ES, Kim SH, Kim YJ, Lee JS, Chung JH (2019) Increased CD3+ T cells with a low FOXP3+/CD8+ T cell ratio can predict anti-PD-1 therapeutic response in non-small cell lung cancer patients. Mod Pathol 32:367–375
Almatroodi SA, McDonald CF, Darby IA, Pouniotis DS (2016) Characterization of M1/M2 tumour-associated macrophages (TAMs) and Th1/Th2 cytokine profiles in patients with NSCLC. Cancer Microenviron 9:1–11
Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, Moy C, Fan Y, Albertyn Z, Huang X, Zhang J, Qiu Y, Platero S, Lorenzi MV, Zudaire E, Yang J, Cheng Y, Xu L, Wu YL (2019) Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun 10:1772
Robinson BW, Morstyn G (1987) Natural killer (NK)-resistant human lung cancer cells are lysed by recombinant interleukin-2-activated NK cells. Cell Immunol 106:215–222
Wang C, Chen Q, Hamajima Y, Sun W, Zheng YQ, Hu XH, Ondrey FG, Lin JZ (2012) Id2 regulates the proliferation of squamous cell carcinoma in vitro via the NF-kappaB/Cyclin D1 pathway. Chin J Cancer 31:430–439
Zhang S, Li M, Ji H, Fang Z (2018) Landscape of transcriptional deregulation in lung cancer. BMC Genom 19:435
Sherman EJ, Mitchell DC, Garner AL (2019) The RNA-binding protein SART3 promotes miR-34a biogenesis and G1 cell cycle arrest in lung cancer cells. J Biol Chem 294:17188–17196
Mezquita L, Auclin E, Ferrara R, Charrier M, Remon J, Planchard D, Ponce S, Ares LP, Leroy L, Audigier-Valette C, Felip E, Zeron-Medina J, Garrido P, Brosseau S, Zalcman G, Mazieres J, Caramela C, Lahmar J, Adam J, Chaput N, Soria JC, Besse B (2018) Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non-small cell lung cancer. JAMA Oncol 4:351–357
Sellers K, Allen TD, Bousamra M 2nd, Tan J, Mendez-Lucas A, Lin W, Bah N, Chernyavskaya Y, MacRae JI, Higashi RM, Lane AN, Fan TW, Yuneva MO (2019) Metabolic reprogramming and Notch activity distinguish between non-small cell lung cancer subtypes. Br J Cancer 121:51–64
Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, Samaniego-Castruita D, Singh D, Seumois G, Alzetani A, Woo E, Friedmann PS, King EV, Thomas GJ, Sanchez-Elsner T, Vijayanand P, Ottensmeier CH (2017) Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol 18:940–950
Osoegawa A, Yoshino I, Tanaka S, Sugio K, Kameyama T, Yamaguchi M, Maehara Y (2004) Regulation of p27 by S-phase kinase-associated protein 2 is associated with aggressiveness in non-small-cell lung cancer. J Clin Oncol 22:4165–4173
Niemira M, Collin F, Szalkowska A, Bielska A, Chwialkowska K, Reszec J, Niklinski J, Kwasniewski M, Kretowski A (2019) Molecular signature of subtypes of non-small-cell lung cancer by large-scale transcriptional profiling: identification of key modules and genes by weighted gene co-expression network analysis (WGCNA). Cancers (Basel) 12
Anderson K, Ryan N, Volpedo G, Varikuti S, Satoskar AR, Oghumu S (2019) Immune suppression mediated by STAT4 deficiency promotes lymphatic metastasis in HNSCC. Front Immunol 10:3095
Karyampudi L, Lamichhane P, Scheid AD, Kalli KR, Shreeder B, Krempski JW, Behrens MD, Knutson KL (2014) Accumulation of memory precursor CD8 T cells in regressing tumors following combination therapy with vaccine and anti-PD-1 antibody. Cancer Res 74:2974–2985
Jian M, Yunjia Z, Zhiying D, Yanduo J, Guocheng J (2019) Interleukin 7 receptor activates PI3K/Akt/mTOR signaling pathway via downregulation of Beclin-1 in lung cancer. Mol Carcinog 58:358–365
Loke P, Allison JP (2003) PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci USA 100:5336–5341
D’Alterio C, Buoncervello M, Ierano C, Napolitano M, Portella L, Rea G, Barbieri A, Luciano A, Scognamiglio G, Tatangelo F, Anniciello AM, Monaco M, Cavalcanti E, Maiolino P, Romagnoli G, Arra C, Botti G, Gabriele L, Scala S (2019) Targeting CXCR4 potentiates anti-PD-1 efficacy modifying the tumor microenvironment and inhibiting neoplastic PD-1. J Exp Clin Cancer Res 38:432
Funding
This work was supported by the Natural Science Foundation of Shanghai (19ZR1449700) and Shanghai Chest Hospital-Collaborative Innovation Project (YJXT20190204).
Author information
Authors and Affiliations
Contributions
Runbo Zhong (RZ) and Yunbin Zhang (YZ) conducted the single-cell sequencing and bioinformatics analysis. Dongfang Chen (DC) collected the samples and Shuhui Cao (SC) collected the clinical data. Baohui Han (BH) and Hua Zhong (HZ) designed the study and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The ethics committee of Shanghai Chest Hospital approved our study, and the patient provided written inform consent.
Consent for publication
Informed consent for publication of the manuscript was obtained from all authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhong, R., Zhang, Y., Chen, D. et al. Single-cell RNA sequencing reveals cellular and molecular immune profile in a Pembrolizumab-responsive PD-L1-negative lung cancer patient. Cancer Immunol Immunother 70, 2261–2274 (2021). https://doi.org/10.1007/s00262-021-02848-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-021-02848-0