Abstract
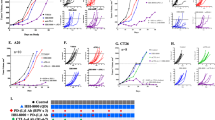
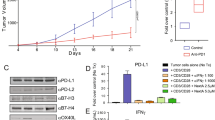
Curaxins are small molecules that bind genomic DNA and interfere with DNA-histone interactions leading to the loss of histones and decondensation of chromatin. We named this phenomenon ‘chromatin damage’. Curaxins demonstrated anti-cancer activity in multiple pre-clinical tumor models. Here, we present data which reveals, for the first time, a role for the immune system in the anti-cancer effects of curaxins. Using the lead curaxin, CBL0137, we observed elevated expression of several group of genes in CBL0137-treated tumor cells including interferon sensitive genes, MHC molecules, some embryo-specific antigens suggesting that CBL0137 increases tumor cell immunogenicity and improves recognition of tumor cells by the immune system. In support of this, we found that the anti-tumor activity of CBL0137 was reduced in immune deficient SCID mice when compared to immune competent mice. Anti-tumor activity of CBL0137 was abrogated in CD8+ T cell depleted mice but only partially lost when natural killer or CD4+ T cells were depleted. Further support for a key role for the immune system in the anti-tumor activity of CBL0137 is evidenced by an increased antigen-specific effector CD8+ T cell and NK cell response, and an increased ratio of effector T cells to Tregs in the tumor and spleen. CBL0137 also elevated the number of CXCR3-expressing CTLs in the tumor and the level of interferon-γ-inducible protein 10 (IP-10) in serum, suggesting IP-10/CXCR3 controls CBL0137-elicited recruitment of effector CTLs to tumors. Our collective data underscores a previously unrecognized role for both innate and adaptive immunity in the anti-tumor activity of curaxins.






Similar content being viewed by others
Data availability
All materials are available upon request.
References
Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD (2017) DNA damage and repair biomarkers of immunotherapy response. Cancer Discov 7(7):675–693. https://doi.org/10.1158/2159-8290.CD-17-0226
Klinakis A, Karagiannis D, Rampias T (2020) Targeting DNA repair in cancer: current state and novel approaches. Cell Mol Life Sci 77(4):677–703. https://doi.org/10.1007/s00018-019-03299-8
Souliotis VL, Vlachogiannis NI, Pappa M, Argyriou A, Ntouros PA, Sfikakis PP (2019) DNA damage response and oxidative stress in systemic autoimmunity. Int J Mol Sci. https://doi.org/10.3390/ijms21010055
Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339(6121):786–791. https://doi.org/10.1126/science.1232458
Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V (2013) cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature 498(7454):380–384. https://doi.org/10.1038/nature12306
Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V (2014) Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J 33(24):2937–2946. https://doi.org/10.15252/embj.201488726
Li A, Yi M, Qin S, Song Y, Chu Q, Wu K (2019) Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J Hematol Oncol 12(1):35. https://doi.org/10.1186/s13045-019-0721-x
Jiang M, Chen P, Wang L, Li W, Chen B, Liu Y, Wang H, Zhao S, Ye L, He Y, Zhou C (2020) cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol 13(1):81. https://doi.org/10.1186/s13045-020-00916-z
Wan D, Jiang W, Hao J (2020) Research advances in how the cGAS-STING pathway controls the cellular inflammatory response. Front Immunol 11:615. https://doi.org/10.3389/fimmu.2020.00615
Gasparian AV, Burkhart CA, Purmal AA, Brodsky L, Pal M, Saranadasa M, Bosykh DA, Commane M, Guryanova OA, Pal S, Safina A, Sviridov S, Koman IE, Veith J, Komar AA, Gudkov AV, Gurova KV (2011) Curaxins: anticancer compounds that simultaneously suppress NF-kappaB and activate p53 by targeting FACT. Sci Transl Med 3(95):95ra74. https://doi.org/10.1126/scitranslmed.3002530
Koman IE, Commane M, Paszkiewicz G, Hoonjan B, Pal S, Safina A, Toshkov I, Purmal AA, Wang D, Liu S, Morrison C, Gudkov AV, Gurova KV (2012) Targeting FACT complex suppresses mammary tumorigenesis in Her2/neu transgenic mice. Cancer Prev Res 5(8):1025–1035. https://doi.org/10.1158/1940-6207.CAPR-11-0529
Carter DR, Murray J, Cheung BB, Gamble L, Koach J, Tsang J, Sutton S, Kalla H, Syed S, Gifford AJ, Issaeva N, Biktasova A, Atmadibrata B, Sun Y, Sokolowski N, Ling D, Kim PY, Webber H, Clark A, Ruhle M, Liu B, Oberthuer A, Fischer M, Byrne J, Saletta F, le Thwe M, Purmal A, Haderski G, Burkhart C, Speleman F, De Preter K, Beckers A, Ziegler DS, Liu T, Gurova KV, Gudkov AV, Norris MD, Haber M, Marshall GM (2015) Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci Transl Med 7(312):312ra176. https://doi.org/10.1126/scitranslmed.aab1803
Burkhart C, Fleyshman D, Kohrn R, Commane M, Garrigan J, Kurbatov V, Toshkov I, Ramachandran R, Martello L, Gurova KV (2014) Curaxin CBL0137 eradicates drug resistant cancer stem cells and potentiates efficacy of gemcitabine in preclinical models of pancreatic cancer. Oncotarget 5(22):11038–11053. https://doi.org/10.18632/oncotarget.2701
Dermawan JK, Hitomi M, Silver DJ, Wu Q, Sandlesh P, Sloan AE, Purmal AA, Gurova KV, Rich JN, Lathia JD, Stark GR, Venere M (2016) Pharmacological targeting of the histone chaperone complex FACT preferentially eliminates glioblastoma stem cells and prolongs survival in preclinical models. Cancer Res 76(8):2432–2442. https://doi.org/10.1158/0008-5472.CAN-15-2162
Barone TA, Burkhart CA, Safina A, Haderski G, Gurova KV, Purmal AA, Gudkov AV, Plunkett RJ (2017) Anticancer drug candidate CBL0137, which inhibits histone chaperone FACT, is efficacious in preclinical orthotopic models of temozolomide-responsive and -resistant glioblastoma. Neuro Oncol 19(2):186–196. https://doi.org/10.1093/neuonc/now141
Leonova K, Safina A, Nesher E, Sandlesh P, Pratt R, Burkhart C, Lipchick B, Gitlin I, Frangou C, Koman I, Wang J, Kirsanov K, Yakubovskaya MG, Gudkov AV, Gurova K (2018) TRAIN (transcription of repeats activates interferon) in response to chromatin destabilization induced by small molecules in mammalian cells. Elife. https://doi.org/10.7554/eLife.30842
Safina A, Cheney P, Pal M, Brodsky L, Ivanov A, Kirsanov K, Lesovaya E, Naberezhnov D, Nesher E, Koman I, Wang D, Wang J, Yakubovskaya M, Winkler D, Gurova K (2017) FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res 45(4):1925–1945. https://doi.org/10.1093/nar/gkw1366
Lock R, Carol H, Maris JM, Kolb EA, Gorlick R, Reynolds CP, Kang MH, Keir ST, Wu J, Purmal A, Gudkov A, Kurmashev D, Kurmasheva RT, Houghton PJ, Smith MA (2017) Initial testing (stage 1) of the curaxin CBL0137 by the pediatric preclinical testing program. Pediatr Blood Cancer. https://doi.org/10.1002/pbc.26263
Somers K, Kosciolek A, Bongers A, El-Ayoubi A, Karsa M, Mayoh C, Wadham C, Middlemiss S, Neznanov N, Kees UR, Lock RB, Gudkov A, Sutton R, Gurova K, Haber M, Norris MD, Henderson MJ (2020) Potent antileukemic activity of curaxin CBL0137 against MLL-rearranged leukemia. Int J Cancer 146(7):1902–1916. https://doi.org/10.1002/ijc.32582
Matsuzaki J, Tsuji T, Luescher IF, Shiku H, Mineno J, Okamoto S, Old LJ, Shrikant P, Gnjatic S, Odunsi K (2015) Direct tumor recognition by a human CD4(+) T-cell subset potently mediates tumor growth inhibition and orchestrates anti-tumor immune responses. Sci Rep 5:14896. https://doi.org/10.1038/srep14896
Nesher E, Safina A, Aljahdali I, Portwood S, Wang ES, Koman I, Wang J, Gurova KV (2018) Role of chromatin damage and chromatin trapping of FACT in mediating the anticancer cytotoxicity of DNA-binding small-molecule drugs. Cancer Res 78(6):1431–1443. https://doi.org/10.1158/0008-5472.CAN-17-2690
Chang HW, Valieva ME, Safina A, Chereji RV, Wang J, Kulaeva OI, Morozov AV, Kirpichnikov MP, Feofanov AV, Gurova KV, Studitsky VM (2018) Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci Adv 4(11):eaav2131. https://doi.org/10.1126/sciadv.aav2131
Fleyshman D, Prendergast L, Safina A, Paszkiewicz G, Commane M, Morgan K, Attwood K, Gurova K (2017) Level of FACT defines the transcriptional landscape and aggressive phenotype of breast cancer cells. Oncotarget 8(13):20525–20542. https://doi.org/10.18632/oncotarget.15656
Kantidze OL, Luzhin AV, Nizovtseva EV, Safina A, Valieva ME, Golov AK, Velichko AK, Lyubitelev AV, Feofanov AV, Gurova KV, Studitsky VM, Razin SV (2019) The anti-cancer drugs curaxins target spatial genome organization. Nat Commun 10(1):1441. https://doi.org/10.1038/s41467-019-09500-7
Grosso JF, Jure-Kunkel MN (2013) CTLA-4 blockade in tumor models: an overview of preclinical and translational research. Cancer Immun 13:5
Stewart R, Morrow M, Hammond SA, Mulgrew K, Marcus D, Poon E, Watkins A, Mullins S, Chodorge M, Andrews J, Bannister D, Dick E, Crawford N, Parmentier J, Alimzhanov M, Babcook JS, Foltz IN, Buchanan A, Bedian V, Wilkinson RW, McCourt M (2015) Identification and characterization of MEDI4736, an antagonistic anti-PD-L1 monoclonal antibody. Cancer Immunol Res 3(9):1052–1062. https://doi.org/10.1158/2326-6066.CIR-14-0191
Wang M, Chen PW, Bronte V, Rosenberg SA, Restifo NP (1995) Anti-tumor activity of cytotoxic T lymphocytes elicited with recombinant and synthetic forms of a model tumor-associated antigen. J Immunother Emphas Tumor Immunol 18(3):139–146. https://doi.org/10.1097/00002371-199510000-00001
Maimela NR, Liu S, Zhang Y (2019) Fates of CD8+ T cells in tumor microenvironment. Comput Struct Biotechnol J 17:1–13. https://doi.org/10.1016/j.csbj.2018.11.004
Huang AY, Gulden PH, Woods AS, Thomas MC, Tong CD, Wang W, Engelhard VH, Pasternack G, Cotter R, Hunt D, Pardoll DM, Jaffee EM (1996) The immunodominant major histocompatibility complex class I-restricted antigen of a murine colon tumor derives from an endogenous retroviral gene product. Proc Natl Acad Sci USA 93(18):9730–9735. https://doi.org/10.1073/pnas.93.18.9730
de Araujo-Souza PS, Hanschke SC, Viola JP (2015) Epigenetic control of interferon-gamma expression in CD8 T cells. J Immunol Res 2015:849573. https://doi.org/10.1155/2015/849573
Groom JR, Luster AD (2011) CXCR3 in T cell function. Exp Cell Res 317(5):620–631. https://doi.org/10.1016/j.yexcr.2010.12.017
Ostroumov D, Fekete-Drimusz N, Saborowski M, Kuhnel F, Woller N (2018) CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell Mol Life Sci 75(4):689–713. https://doi.org/10.1007/s00018-017-2686-7
Kalathil SG, Hutson A, Barbi J, Iyer R, Thanavala Y (2019) Augmentation of IFN-gamma+ CD8+ T cell responses correlates with survival of HCC patients on sorafenib therapy. JCI Insight. https://doi.org/10.1172/jci.insight.130116
Perdicchio M, Cornelissen LA, Streng-Ouwehand I, Engels S, Verstege MI, Boon L, Geerts D, van Kooyk Y, Unger WW (2016) Tumor sialylation impedes T cell mediated anti-tumor responses while promoting tumor associated-regulatory T cells. Oncotarget 7(8):8771–8782. https://doi.org/10.18632/oncotarget.6822
Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W (2018) CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 18(10):635–647. https://doi.org/10.1038/s41577-018-0044-0
Ben-Shmuel A, Biber G, Barda-Saad M (2020) Unleashing natural killer cells in the tumor microenvironment-the next generation of immunotherapy? Front Immunol 11:275. https://doi.org/10.3389/fimmu.2020.00275
Schoenborn JR, Wilson CB (2007) Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol 96:41–101. https://doi.org/10.1016/S0065-2776(07)96002-2
Simonetta F, Pradier A, Roosnek E (2016) T-bet and eomesodermin in NK cell development, maturation, and function. Front Immunol 7:241. https://doi.org/10.3389/fimmu.2016.00241
Werneck MB, Lugo-Villarino G, Hwang ES, Cantor H, Glimcher LH (2008) T-bet plays a key role in NK-mediated control of melanoma metastatic disease. J Immunol 180(12):8004–8010. https://doi.org/10.4049/jimmunol.180.12.8004
Kallies A, Good-Jacobson KL (2017) Transcription factor T-bet orchestrates lineage development and function in the immune system. Trends Immunol 38(4):287–297. https://doi.org/10.1016/j.it.2017.02.003
Mocellin S, Wang E, Marincola FM (2001) Cytokines and immune response in the tumor microenvironment. J Immunother 24(5):392–407
Griffith JW, Sokol CL, Luster AD (2014) Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 32:659–702. https://doi.org/10.1146/annurev-immunol-032713-120145
Pang B, Qiao X, Janssen L, Velds A, Groothuis T, Kerkhoven R, Nieuwland M, Ovaa H, Rottenberg S, van Tellingen O, Janssen J, Huijgens P, Zwart W, Neefjes J (2013) Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun 4:1908. https://doi.org/10.1038/ncomms2921
Yang F, Kemp CJ, Henikoff S (2013) Doxorubicin enhances nucleosome turnover around promoters. Curr Biol 23(9):782–787. https://doi.org/10.1016/j.cub.2013.03.043
Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remedios C, Fend L, Hannani D, Aymeric L, Ma Y, Niso-Santano M, Kepp O, Schultze JL, Tuting T, Belardelli F, Bracci L, La Sorsa V, Ziccheddu G, Sestili P, Urbani F, Delorenzi M, Lacroix-Triki M, Quidville V, Conforti R, Spano JP, Pusztai L, Poirier-Colame V, Delaloge S, Penault-Llorca F, Ladoire S, Arnould L, Cyrta J, Dessoliers MC, Eggermont A, Bianchi ME, Pittet M, Engblom C, Pfirschke C, Preville X, Uze G, Schreiber RD, Chow MT, Smyth MJ, Proietti E, Andre F, Kroemer G, Zitvogel L (2014) Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med 20(11):1301–1309. https://doi.org/10.1038/nm.3708
Jin MZ, Xia BR, Xu Y, Jin WL (2018) Curaxin CBL0137 exerts anticancer activity via diverse mechanisms. Front Oncol 8:598. https://doi.org/10.3389/fonc.2018.00598
Luster AD, Unkeless JC, Ravetch JV (1985) Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315(6021):672–676. https://doi.org/10.1038/315672a0
Warhurst AC, Hopkins SJ, Warhurst G (1998) Interferon gamma induces differential upregulation of alpha and beta chemokine secretion in colonic epithelial cell lines. Gut 42(2):208–213. https://doi.org/10.1136/gut.42.2.208
Matsukura S, Kokubu F, Kuga H, Kawaguchi M, Ieki K, Odaka M, Suzuki S, Watanabe S, Takeuchi H, Adachi M, Stellato C, Schleimer RP (2003) Differential regulation of eotaxin expression by IFN-gamma in airway epithelial cells. J Allergy Clin Immunol 111(6):1337–1344. https://doi.org/10.1067/mai.2003.1513
Liao F, Rabin RL, Yannelli JR, Koniaris LG, Vanguri P, Farber JM (1995) Human Mig chemokine: biochemical and functional characterization. J Exp Med 182(5):1301–1314. https://doi.org/10.1084/jem.182.5.1301
Taub DD, Lloyd AR, Conlon K, Wang JM, Ortaldo JR, Harada A, Matsushima K, Kelvin DJ, Oppenheim JJ (1993) Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med 177(6):1809–1814. https://doi.org/10.1084/jem.177.6.1809
Damsker JM, Hansen AM, Caspi RR (2010) Th1 and Th17 cells: adversaries and collaborators. Ann N Y Acad Sci 1183:211–221. https://doi.org/10.1111/j.1749-6632.2009.05133.x
Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ (2001) T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA 98(26):15137–15142. https://doi.org/10.1073/pnas.261570598
Di Mitri D, Toso A, Alimonti A (2015) Molecular pathways: targeting tumor-infiltrating myeloid-derived suppressor cells for cancer therapy. Clin Cancer Res 21(14):3108–3112. https://doi.org/10.1158/1078-0432.CCR-14-2261
Zeligs KP, Neuman MK, Annunziata CM (2016) Molecular pathways: the balance between cancer and the immune system challenges the therapeutic specificity of targeting nuclear factor-kappaB signaling for cancer treatment. Clin Cancer Res 22(17):4302–4308. https://doi.org/10.1158/1078-0432.CCR-15-1374
Mancino A, Lawrence T (2010) Nuclear factor-kappaB and tumor-associated macrophages. Clin Cancer Res 16(3):784–789. https://doi.org/10.1158/1078-0432.CCR-09-1015
Funding
This study was funded in part by Roswell Park Alliance Foundation (KG), National Cancer Institute (NCI) grant R01CA197967 (KG), Incuron, LLC (KG), and NCI Cancer Center Support Grant P30CA16056 (FICSR).
Author information
Authors and Affiliations
Contributions
MC: performed most of experiments, wrote the manuscript, CMB: design of experiments, analyses of data, editing of manuscript, LB: performed part of mouse experiments, edited manuscript, AP: performed NY-ESO-1-specific ELISPOT assay, SKP: analyzed data, edited manuscript, JM: performed NY-ESO-1-specific ELISPOT assay, edited manuscript, AO: edited manuscript, AG: edited manuscript, AKS: design of experiments, analyses of data, editing of manuscript, ER: design of experiments, analyses of data, editing of manuscript, KG: design of experiments, analyses of data, editing of manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
K. Gurova is co-inventor of curaxins and recipient of research grants and consulting payments from Incuron, LLC.
Ethical approval
The protocols of all experiments with mice in this study were approved by the IACUC at Roswell Park Comprehensive Cancer Center.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, M., Brackett, C.M., Burdelya, L.G. et al. Stimulation of an anti-tumor immune response with “chromatin-damaging” therapy. Cancer Immunol Immunother 70, 2073–2086 (2021). https://doi.org/10.1007/s00262-020-02846-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-020-02846-8