Abstract
Immune-mediated rejection of human cancer is a relatively rare but well-documented phenomenon. Its rate of occurrence progressively increases from the occasional observation of spontaneous regressions to the high rate of complete remissions observed in response to effective treatments. For two decades, our group has focused its interest in understanding this phenomenon by studying humans following an inductive approach. Sticking to a sequential logic, we dissected the phenomenon by studying to the best of our capability both peripheral and tumor samples and reached the conclusion that immune-mediated cancer rejection is a facet of autoimmunity where the target tissue is the cancer itself. As we are currently defining the strategy to effectively identify the mechanisms leading in individual patients to rejection of their own tumors, we considered useful to summarize the thought process that guided us to our own interpretation of the mechanisms of immune responsiveness.
Similar content being viewed by others
The molecular basis of T cell-mediated immune recognition and the self–non-self preposition
The first evidence that T cells can recognize cancer was based on the description by Wolfel et al. [1] of tumor infiltrating lymphocytes (TILs) that could kill autologous tumor cells following a human leukocyte antigen (HLA)-restricted pattern and by Kawakami et al. [2] who characterized the requirements for HLA restriction. In 1991, the molecular characterization of the first tumor antigen (TA) recognized by T cells was done by Van der Bruggen et al. [3]. This discovery, followed by the identification or several other TA-specific molecular targets of T cells [4, 5], provided the opportunity for tumor-specific immunotherapeutic intervention [6]. Contemporaneously, others identified naturally occurring antibodies directed against TAs of which several were also recognized by T cells [7].
One category of TAs included melanoma differentiation antigens (MDA) whose expression is shared by melanoma cells and normal epithelial melanocytes. Another category includes proteins whose expression was associated with the progressive de-methylation of cancer cells; as these non-mutated proteins were expressed selectively by germ line cells in the testes and cancer cells, they were named cancer testing antigens (CTA).
The identification of MDAs and CTAs and their respective epitopes associated with specific HLA molecules spurred interest in using these molecules and their derivatives for TA-specific active immunization [8]. An increasing body of literature has more recently focused on TA derived from infectious agents such as human papilloma virus or immunogenic mutated proteins whose expression may or may not be shared by different patients’ tumors depending upon the frequency of a given mutation. These TAs may represent a distinct and important facet of T cell-mediated recognition of tumors. Indeed, recent work suggests that immunization against viral antigens may be particularly promising [9]. The current viewpoint represents a retrospective evaluation of what we learnt predominantly studying MDA-specific vaccinations; we believe that part of the learning may apply to a broader range of phenomena related to T cell-based immunotherapy of cancer while in other cases, more immunogenic non-self TA may stand on their own. Future work should include of course these approaches for which at the moment there is relatively less clinical experience.
Epitope-specific anti-cancer vaccines as a model to understand immune rejection
Despite the large number of trials vaccinating patients with TAs, results have been in general disappointing [8, 10, 11], although in a minority of cases, vaccines may prolong the frequency of clinical responses and/or survival [12–14]. One of them is now the first anti-cancer vaccine licensed by the Food and Drug Administration. Independent of their clinical potential, epitope-specific anti-cancer vaccines, however, provided to immunologists the unprecedented opportunity to study in relatively controlled settings how the human immune system functions. Thus, it could be tested whether vaccine-induced circulating TA-specific T cells could reach the tumor site and recognize their target [15]; the use of tetrameric HLA complexes [16] made it possible to characterize ex vivo the functional properties of vaccine-induced T cells [17, 18].
The paradoxical co-existence of tumor-specific T cells (in the circulation and in the tumor) and their target
Anti-cancer vaccination efforts consistently elicited expansion of TA-specific memory T cells in the peripheral circulation, but this was rarely accompanied by cancer regression [19], suggesting that tumor rejection is a complex phenomenon that goes beyond the recognition of target cells by T cells [20]. Several questions emerged: do vaccine-induced T cells reach the tumor site? Does the tumor microenvironment provide sufficient co-stimulation to activate otherwise quiescent T cells [21]? How do the evolving phenotypes of cancer cells escape potentially effective immune responses? Most intriguingly, are tumor-changing phenotypes responsible for cancer cell escape from recognition by an adequately activated immune response? Alternatively, are the immune responses elicited by vaccines insufficient to destroy cancer cells that are otherwise adequately expressing target molecules [21]?
The quiescent status of vaccine-induced T cell
Lee et al. [22] documented a status of anergy of spontaneously occurring MDA-specific circulating T cells. Further work demonstrated that, beyond stage II, patients with breast and colon cancer or melanoma suffer a profound depression of innate immune responses. This immune suppression is exemplified by reduced production of IFN-γ in response to TA-specific stimulation ex vivo [22] and reduced inducible levels of signal transducer and activator of transcription (STAT)-1 phosphorylation in circulating immune cells following ex vivo stimulation with type I interferon (IFN) [23, 24]. Interestingly, it was observed that although patients with cancer experience a significantly reduced responsiveness to IFN-α stimulation compared with healthy individuals, such depression occurs within a big range of values with some patients demonstrating normal response to stimulation also at a later stage of disease. This suggests that some aspects of the host’s or the individual tumor biology may differ dramatically among different cancer-bearing subjects. Similar findings were later reported by others, who observed in patients with cancer the same depression in STAT-1 phosphorylation following stimulation with IL-2 [25]. It also became apparent that spontaneously occurring anti-cancer immune responses in patients with metastatic melanoma displayed a memory phenotype [26, 27] providing evidence for in vivo priming of effector T cells by the cancer-bearing status [26].
While the work of Lee and other groups focused on spontaneously occurring TA-specific T cells, others analyzed the type and function of vaccine-elicited T cells documenting that naturally occurring TA-specific immune responses could be consistently enhanced by immunization [28, 29]. Contrary to naturally occurring TA-specific T cells incapable of expressing IFN-γ in response to cognate stimulation [22], vaccine-induced T cells were able to produce IFN-γ ex vivo [17]. However, they lacked cytotoxic function that could be recovered by in vitro activation with TA-specific stimulation in the presence of recombinant interleukin (IL)-2. We defined these incompletely anergic vaccine-induced T cells as “quiescent” [18]. This quiescent phenotype was reminiscent of the contraction phase of effector immune responses described by Kaech [30] according to the linear model of T cell activation. According to this model, following a time-delimited stimulation, T cells expand and become activated during the first week, acquiring also cytotoxic properties. In the following 2–3 weeks, CD8 T cells contract in number and, while maintaining responsiveness to antigen-specific stimulation, loose cytotoxic function. This model well approximates the kinetics of TA-specific immunization, which is provided at intervals of 1–3 weeks, and T cell immune responses are generally tested weeks after immunization. This observation raised the question: assuming that vaccine-induced T cells reach the tumor site, could the tumor in steady state conditions or during immune therapy provide a milieu conducive to their reactivation comparable to that simulated in vitro by re-stimulation with TA in the presence of IL-2 [18]? While most remained interested in the afferent aspects of vaccination and continued to develop increasingly more sophisticated strategies aimed at inducing qualitatively and quantitatively better immune responses, we turned our interest toward the efferent loop to understand the requirements for their activation in the target organ. This was based on the assumption that circulating vaccine-induced T cells were fully functional according to the physiology and the dynamics of T cell activation in response to a time-limited stimulus such as a vaccine [21]. It should be clarified that the afferent aspect of immunization include mode of action studies and the pharmacodynamics that are integral part of a rational development of immune therapies aiming at inducing specific immune responses while the efferent aspect includes the study of those variables that determine the effects of vaccine-induced immune responses after their induction. It should also be emphasized that the study of the afferent loop of tumor vaccines remains important for drug developers aiming to understand the mechanism of action of their products; many basic questions remain unsolved (which antigen format, which adjuvant, which route of administration, which dosing schedule, etc.). Thus, although drug developers made in the past the mistake of concentrating mostly on the afferent loop neglecting the efferent one, we should not make the reverse mistake now but rather combine the two approaches into an integrated and systematic analysis of all potential variables that could decode the algorithm governing immune responsiveness.
The limited role of tumor immune escape as predictor of immune responsiveness
Evolution under selection implies that forceful negative pressure is imposed upon malleable phenotypes. To explain the co-existence of a cognate immune response against cancer with its concurrent growth, immune selection is often invoked [31, 32]. This concept revolves around the demonstration in experimental and clinical models that immune suppression is associated with higher incidence of spontaneous cancers [33, 34]. Recently, this concept is enjoying broader clinical recognition as the relevance of immune infiltrates on the natural history of cancer is becoming increasingly appreciated [35, 36]. To explain why tumors grow in the presence of an active immune response, the work of several groups including ours has described and characterized several potential mechanisms by which tumors could escape immune recognition [20, 32, 37–39]. However, these observations refer to conditions when tumors do not undergo immunotherapy and a balance is stricken between the host’s immune response and cancer growth. Therefore, it is likely that immune escape, also referred to as immune editing [40], may play a significant role at the onset and throughout the natural history of cancer. However, to establish causation in the determinism of responsiveness to immunotherapy, level of TA and TA presentation should be assessed before treatment and a predictive relationship should be demonstrated on the likelihood to respond to the respective treatment. In fact, the balance stricken between the host’s immune response and its cancer in natural conditions is shaken by powerful immune responses observable during an acute inflammatory process [41]. Thus, the described mechanism that could explain reduced recognition of tumor cells in natural conditions may not apply to the lack of tumor responsiveness to effective immunotherapy when the balanced is switched suddenly in favor of the host, and cancer cells had no time to adapt to the novel condition. As later described, direct human observations of tumors performed before treatment did not demonstrate that the level of TA expression predicts response to therapy. Those studies suggested that the lack of response to immunotherapy is due to limited activation of immune responses within the target organ rather than the loss of antigenic properties by cancer cells. Loss of TA expression appears only after treatment in recurring lesions following partially successful therapy that did not completely eradicate all cancer cells [20] (Fig. 1).
Extremes in the interpretation of the mechanisms leading to tumor rejection and their impact on the relevance of the heterogeneity of tumor cells in determining their escape from immune recognition. a In the simplest scenario, adaptive immune responses, whether mediated by cytotoxic T cells (CTLs) or antibody-mediated cytotoxicity, play a prominent role in eliminating cancer cells expressing the appropriate epitope; in a cancer population with heterogeneous expression of target epitope due to loss of antigen or antigen processing and presentation defects, only the epitope expressing cancer cells would be eliminated leaving the other ones alive (b). c An alternative scenario suggests that the effector/target cell complex may deliver pro-inflammatory signals due to the release of cytokines or damage-associated molecular patterns (DAMPs; in the figure, in red are those associated with T cell effector function, in blue those associated with antibody-mediated cytotoxicity). These, in turn, attract and activate innate immune effectors such as natural killer (NK) cells or macrophages which can exert cytotoxic functions on cancer cells independent of epitope expression. This, in turn, may lead to a broader clearance of cancer cells leaving only macrophages in charge of tissue repair (d). Which of these scenarios more closely represent human reality remains to be determined
On the necessity to study the tumor microenvironment and the serial biopsy strategy
Direct observation of the tumor microenvironment may obviate for the need of hypotheses [42, 43]. As TSD is tissue-specific, it is best to study the target organ: the cancer itself. There is a dissonance between the current emphasis on the characterization of circulating immune cells and the indirect role that they play in the cancer microenvironment. The approach lacks appreciation for the heterogeneity of individual tumors and their dynamic instability [44]. Thus, to understand human cancer immune responsiveness, tumors should be studied in steady state conditions to document patient-specific idiosyncrasies and during therapy to discover phenomena relevant to TSD. It should be kept in mind that immune manipulation of the host may not only effect the function of immune cells but, directly or indirectly, the function also of cancer cells or other normal cells within the tumor microenvironment. It is only during those dynamic moments that the mechanisms relevant to rejection will emerge; only at those moments, modulation of HLA expression [45–48] or changes in cytokines-dependent modulation of the immune microenvironment become apparent as well as exemplified by the dual role played by IL-10 that will be discussed later [49]. Thus, we conclude that tumor-related TSD could be effectively studied by catching the switch from a chronic to an acute inflammatory process by serially studying individual tumors before, during, and after treatment [50].
It soon became apparent, however, the classical approaches to the study of tumors were not suitable. Most studies utilized excisional biopsies, which provide optimal quantity of tissue and allow its morphological assessment. However, because the tumor is removed, serial sampling of the same lesion for dynamic studies cannot be done. Moreover, no direct correlation can be performed between the findings obtained studying the excised material and the response to immunotherapy of concomitant metastases left in the patient. The assumption that removed tumors are representative of those left in the patient as targets of therapy was soon proved inaccurate; the study of synchronous metastases demonstrated significant variability among most markers evaluated [51]. We, therefore, applied in subsequent studies minimally invasive biopsies such as fine needle aspirate (FNA) biopsies that allowed serial sampling of the same lesions [50].
Localization of vaccine-induced T cells at tumor site may be necessary but it is not sufficient for tumor regression
Localization within tumors of indium111-labeled adoptively transferred TILs is necessary for clinical response [52]; however, TIL localization is not sufficient, and several lesions demonstrating uptake do not respond to therapy.
Since TIL localization in the target organ is necessary for tumor rejection, we tested whether vaccine-induced T cells reached the tumor site. Kammula et al. [15] applied serial FNAs to melanoma metastases, comparing the expression of interferon (IFN)-γ before and during vaccination. This study was performed on lesions whose cancer cells did express sufficient amount of the relevant HLA allele to exclude escape from T cell recognition; expression of the MDA target of the vaccination was monitored in parallel. Furthermore, localization of vaccine-specific T cells was monitored with tetrameric HLA/peptide complexes. This study clearly documented a significant enhancement in IFN-γ expression in tumors following vaccination, and the level of expression of this cytokine correlated with the level of expression of the relevant MDA. Thus, vaccine-induced T cell do reach the tumor site, interact with tumor cells by producing IFN-γ and, therefore, are not anergic. However, this functionally competent localization was not sufficient for tumor rejection as all lesions continued to grow. This finding mirrors the quiescent phenotype of vaccine-induced T cells observed in the circulation that could produce IFN-γ when encountering the relevant TA but cannot exert broader effector functions.
The surprising role of IL-10
Several studies suggest that tumors produce immune regulatory molecules that could suppress the function of T cells. Applying FNA biopsies, we assessed in pre-treatment samples the expression of cytokines with potential regulatory effects [53]. Surprisingly, the expression levels of none of them predicted unresponsiveness; on the contrary, higher levels of IL-10 significantly predicted response. Further analyses demonstrated that the IL-10 was not only expressed at the RNA but also at the protein level by cancer cells. Retrospectively, this observation was not surprising as several other studies had observed that, contrary to the expectations, IL-10 overexpression was a predictor of immune responsiveness. This cytokine probably favors tumor growth in steady state conditions by inhibiting the maturation of antigen presenting cells, but at the same time, sustaining their ability for antigen uptake and simultaneously hampering their migration to draining lymph nodes prepares antigen presenting cells to serve as powerful T cell stimulators loaded with TA when the microenvironmental conditions are altered during immune stimulation that could affect a switch of their phenotype by the presence of immune stimulatory cytokines such as IFN-γ and TNF-α that could induce their maturation in situ [49].
Levels of HLA/TA expression by cancer cells does not predict immune response
It is logical that the success of TA-specific vaccination is predicated upon the level of expression of the target TA by cancer cells. We, therefore, measured by FNA in pre- and post-treatment metastases the expression of TA relevant and irrelevant to the vaccine administered [54]. The analysis included only melanoma metastases whose cancer cells expressed the associated HLA molecule necessary for T cell recognition. The level of expression of the target TA was not predictive of responsiveness. However, we observed that lesions biopsied soon after treatment as they were undergoing a biological response that subsequently led to their complete disappearance dramatically reduced the expression of the TA target of the vaccine. This observation suggests that when immunotherapy works, the first targets of therapy are the cancer cells expressing the relevant TA. The loss of TA expression was not associated with complete disappearance of cancer cells at that time point as other TA expressed specifically by cancer cells such as CTAs were still present, and cancer cells could still be observed in the cytospins. Thus, loss of cancer cells expressing TA targeted by the vaccine preceded the elimination of the complete neoplastic population. Since the metastases subsequently underwent complete disappearance, it became clear that the effect of vaccination was to initiate a self-perpetuating process that continued beyond the initial interaction between vaccine-induced T cells and their targets [20, 55]. The same conclusion was subsequently independently reached by others in the context of CTA-specific anti-cancer vaccination [56].
The advent of whole-genome, hypothesis-generating studies
The dynamic approach to the study of immune responsiveness had, by the end of the 1990s, crushed several myths. Hundreds of modulatory mechanisms that could hamper immune responsiveness could be postulated but such number was too high to be realistically tackled by future studies with a minimalistic approach. Around that time, whole-genome transcriptional assays enter the scene of clinical investigation [57]. The hypothesis-generating strategy well suited our lack of a dominant hypothesis and proved invaluable for the application of an inductive approach aimed at decoding the multifactorial algorithm governing tumor immune responsiveness [41, 43, 50, 55, 58–60]. In particular, we became interested in characterizing two phenomena: first, document the mechanism of tumor rejection by sampling lesions during and after treatment and comparing those that eventually responded to those that did not. Second, dissect the reasons why some but not all tumors respond to identical treatments.
The pre-determinism of immune responsiveness
The first attempt to address at the whole genome level the determinism leading to tumor rejection was an analysis of melanoma metastases biopsied before and after treatment with TA-specific vaccination and concomitant systemic administration of IL-2 [45]. Analysis of pre-treatment biopsies identified a set of genes differentially expressed by lesions that subsequently responded to treatment. Although those early array platforms contained few immunologically relevant genes, the transcripts that differentiated responding from non-responding metastases had predominantly immune function. This observation leads to the conclusion that some metastases were pre-determined to respond to immunotherapy by a pre-existing inflammatory process that although not sufficient to induce spontaneous tumor regression was conducive to immune stimulation. A decade later, we could validate in a small prospective mechanistic study these findings [47]: pre-treatment biopsies in patients with metastatic melanoma who subsequently responded to systemic IL-2 administration displayed a pre-existing pro-inflammatory status. Simultaneously, others identified similar signatures to be predictive of responsiveness in patients with melanoma vaccinated with four TAs plus IL-12 [61], dendritic cells loaded with multiple TAs [62], or melanoma and lung cancer patients receiving a MAGEA3 peptide vaccine [63].
Similarity between cancer rejection and other aspects of immune-mediated tissue-specific destruction (TSD)
In a separate study, we evaluated the mechanism of action of systemic IL-2 administration by sampling melanoma metastases before and during treatment. This study demonstrated that the mechanism of action of IL-2 is an indirect activation of macrophages mediated by a cytokine storm released by circulating IL-2 receptor-bearing cells [64]. The effects at the tumor site were the degrees of magnitude less than at the systemic level. Moreover, the intra-tumor effects were delayed, and the magnitude was dose dependent. This could explain why immune responsiveness has been associated with number of doses received by patients [65]. Because one of the lesions from the six patients studied underwent clinical response, we had the first glimpse at the transcriptional machinery specific for tumor rejection [64]. Surprisingly, several of the transcripts expressed specifically by the responding lesion had been simultaneously described by another group as markers of acute kidney allograft rejection [66]. This was the first suggestion that tumor regression, allograft rejection, and probably other immune destructive processes were facets of a common phenomenon that shared a convergent final pathway.
From the delayed allergy reaction to the immunologic constant of rejection (ICR)
In 1969, Jonas Salk [67] hypothesized that allograft rejection, tumor rejection, and various autoimmune phenomena represented facets of a similar immune-mediated phenomenon that he called the “delayed allergy reaction”. His intuition was validated decades later by work from those who studied with high throughput technology tissue undergoing TSD. This unbiased approach applied to acute allograft rejection, tissues affected by graft versus host disease or flare of autoimmunity, acutely infected organs undergoing clearance of pathogen and tumor rejection during immunotherapy identified a convergent pathway and a group of genes that were always required for TSD to occur. We named this signature “the immunologic constant of rejection (ICR)” [55]. The ICR includes at least 4 functional components: the activation of (1) the IFN-γ/STAT1/IRF-1 pathway [41], (2) immune effector mechanisms, (3) CXCR3, and (4) CCR5 ligand chemokines [68].
How defining the ICR guided the understanding of immune responsiveness
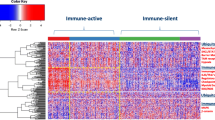
The ICR axiom describes how TSD occurs independent of its cause; in other words, the ICR dictates “how” but not “why” rejection occurs. However, the definition of a necessary pathway clarified the premises for the identification of its causes. It became clear that the lack of rejection of tumors during immunotherapy is due to insufficient stimulation of immune responses at the tumor site rather than to the escape of tumors from a fully activated immune response. Analysis of patients with melanoma experiencing a mixed responses [48] and comparison of tumors responding and non-responding to the same treatment [45, 47] demonstrated that while responding tumors displayed a strongly activated acute inflammatory status, non-responding ones were completely immunologically silent as if the treatment had not reached the target tissue. Moreover, the definition of the ICR leads to the conclusion that genes and pathways activated during TDS are qualitatively similar to those associated with a good prognostic connotation in various cancers [35, 36, 69] and to those predicting immune responsiveness to distinct type of immunotherapy [45, 47, 61, 63, 70]. Thus, a general theory of rejection could be constructed, whereby a progression occurs in the host versus cancer relationship, starting with immune surveillance that slows but does not abrogate tumor growth to a pre-determinism to respond to therapy to a full blown activation of the immune response leading to cancer rejection during treatment. Similar signatures are observed throughout this continuum but the intensity and breath of their activation increases progressively till TDS is reached.
A genetic inference on human cancer immune responsiveness; the role of the genetic background of the host, neoplastic instability, and external factors
Perhaps, the most important contribution offered by the ICR concept in deconvoluting the determinism of tumor rejection is the provision of a road map defined by evidence-based hypotheses. Identification of a specific group of genes, whose activation is necessary for TSD, leads to the analysis of the role of IRF5 polymorphism in melanoma immune responsiveness. This study was based on the premise that, if the ICR applies to autoimmunity and if cancer rejection represents an aspect of autoimmunity, genes relevant to the development of autoimmunity ought to influence the immune biology of cancer. IRF5 plays a significant role in the modulation of several autoimmune diseases including systemic lupus erythematosus (SLE). We recently observed that the same polymorphisms protective against the development of SLE predict non-response to adoptive transfer of TILs (unpublished own data). Similarly, appreciation for the central role played by the CXCR3/CCR5 cluster in mediating TSD leads us to the investigation of its expression by TILs. Integration of germ line, transcriptional, and protein expression data brought to the formulation of a protein prediction model strongly predictive of treatment outcome (unpublished own data). It is important to note that in this study, the expression of individual receptors was not highly predictive of immune responsiveness but the combined over- or underexpression was strongly pointing to the need to look at the biomarkers of immune responsiveness not as individual entities but in accordance with their combined function. Importantly, this second-generation, evidence-based, and hypothesis-driven studies enlightened an underappreciated fault of current bioinformatics approaches applied to the identification of biomarkers of immune responsiveness. This study demonstrated that individual, univariate approaches bear, when examined individually, very little predictive value but the combination of factors integrated according to a logical stream may progressively break the code governing the algorithm of immune responsiveness.
The genetics of the host, the tumor, and the environment
The relative weight played by the host’s genetic background, the somatic alterations acquired during the neoplastic process, and the influence of environmental factors in determining immune responsiveness is unclear. Observations performed during the last two decades strongly suggest that it is a combination of the three that determine immune responsiveness [60]. It is likely that inherited genetic factors may affect the biology of the host or their cancer cells to determine the likelihood of immune responsiveness; to this first checkpoint, the genetics of cancer cells may add its own predisposition to being susceptible to immune attack. Finally, various undetermined variables encompassing the effectiveness of treatment, general status of health and other “hidden” external factors may contribute to the final outcome. Only when all checkpoints are permissive, tumor rejection is observed. Moreover, the genetic weight that the host’s background plays on tumor biology is often underestimated. Tumors from individual patients are close to each other biologically when compared to those from other patients; this may not only be due to their clonal derivation as often assumed [71, 72], but it could also depend upon the innumerable polymorphisms inherited by cancer cells from their host. Thus, the genetic background of the host may affect immune responsiveness, not only by affecting the function of normal immune cells but also by directly affecting the biology of cancers cells. This multistep inference on the mechanism of immune responsiveness may also explain why it is generally easier to predict accurately lack of responsiveness than responsiveness; it is necessary to have a gun to go duck hunting; if no gun is available no duck will be caught; but having a gun is not sufficient as ability to shoot, good visibility, presence of ducks, and many other limiting factors may determine the success of the hunt.
The future challenges and opportunities
In the same manuscript where the delayed allergy reaction was described, Jonas Salk [67] also stated: “The answers pre-exist and are the questions that need to be identified”. We totally agree. The algorithm governing tumor response to immunotherapy is lying in front of us. The recipe to its identification is simple: first, study the tumor together with the peripheral circulation; second, study the genetic background of the host together with the genetics of the tumor; third, apply integrative bioinformatics approaches searching for complex relationships rather than univariate class comparisons. We are confident that this recipe will soon lead to the understanding of tumor immune responsiveness with a caveat: we need to sensitize those who design clinical trials to the need to learn from clinical investigations rather than limiting the testing to the clinical endpoint [73].
Abbreviations
- CTA:
-
Cancer testing antigen
- FNA:
-
Fine needle aspiration biopsy
- HLA:
-
Human leukocyte antigen
- ICR:
-
Immunologic constant of rejection
- IEFs:
-
Immune effector genes
- IFN:
-
Interferon
- IL:
-
Interleukin
- IRF:
-
Interferon regulatory factor
- ISGs:
-
Interferon-stimulated genes
- MDA:
-
Melanoma differentiation antigens
- SLE:
-
Systemic lupus erythematosus
- STAT-1:
-
Signal transducer and activator of transcription
- TIL:
-
Tumor infiltrating lymphocyte
- TA:
-
Tumor antigen
- TSD:
-
Immune-mediated tissue-specific destruction
References
Wolfel T, Klehmann E, Muller C, Schutt KH, Meyer zum Buschenfelde KH, Knuth A (1989) Lysis of human melanoma cells by autologous cytolytic T cell clones (1989) Identification of human histocompatibility leukocyte antigen A2 as a restriction element for three different antigens. J Exp Med 170:797–810
Kawakami Y, Zakut R, Topalian SL, Stotter H, Rosenberg SA (1992) Shared human melanoma antigens. Recognition by tumor-infiltrating lymphocytes in HLA-A2.1-transfected melanomas. J Immunol 148:638–643
van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B et al (1991) A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254:1643–1647
Boon T, Cerottini J-C, Van den Eynde B, van der Bruggen P, Van Pel A (1994) Tumor antigens recognized by T lymphocytes. Annu Rev Immunol 12:337–365
Rosenberg SA, Kawakami Y, Robbins PF, Wang R (1996) Identification of the genes encoding cancer antigens: implications for cancer immunotherapy. Adv Cancer Res 70(145–77):145–177
Rosenberg SA (1997) Cancer vaccines based on the identification of genes encoding cancer regression antigens. Immunol Today 18(4):175–182
Old LJ, Chen YT (1998) New paths in human cancer serology. J Exp Med 187(8):1163–1167
Parmiani G, Castelli C, Dalerba P, Mortarini R, Rivoltini L, Marincola FM et al (2002) Cancer immunotherapy with peptide-based vaccines: what have we achieved? Where are we going? J Natl Cancer Inst 94(11):805–818
van der Burg SH, Melief CJ (2011) Therapeutic vaccination against human papilloma virus induced malignancies. Curr Opin Immunol 23(2):252–257
Rosenberg SA, Yang JC, Restifo NP (2004) Cancer immunotherapy: moving beyond current vaccines. Nat Med 10(9):909–915
Mocellin S, Mandruzzato S, Bronte V, Marincola FM (2004) Correspondence 1: cancer vaccines: pessimism in check. Nat Med 10(12):1278–1279
Schwartzentruber DJ, Lawson DH, Richards JM, Conry RM, Miller DM, Treisman J et al (2011) gp100 Peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med 364(22):2119–2127
Bulloch MN, Elayan MM, Renfroe HR (2011) Sipuleucel-T: a therapeutic cancer vaccine for the treatment of castration- or hormone-refractory prostate cancer. Expert Rev Clin Pharmacol 4(6):685–692
Brichard VG, Lejeune D (2007) GSK’s antigen-specific cancer immunotherapy programme: pilot results leading to phase III clinical development. Vaccine 25(Suppl 2):B61–B71
Kammula US, Lee K-H, Riker A, Wang E, Ohnmacht GA, Rosenberg SA et al (1999) Functional analysis of antigen-specific T lymphocytes by serial measurement of gene expression in peripheral blood mononuclear cells and tumor specimens. J Immunol 163:6867–6879
Altman JD, Moss PA, Goulder PR, Barouch DH, McHeyzer-Williams MG, Bell JI et al (1996) Phenotypic analysis of antigen-specific T lymphocytes [published erratum appears in Science 1998 Jun 19;280(5371):1821] Science 274(5284):94–96
Nielsen M-B, Monsurro’ V, Miguelse S, Wang E, Perez-Diez A, Lee K-H et al (2000) Status of activation of circulating vaccine-elicited CD8+ T cells. J Immunol 165(4):2287–2296
Monsurro’ V, Wang E, Yamano Y, Migueles SA, Panelli MC, Smith K et al (2004) Quiescent phenotype of tumor-specific CD8+ T cells following immunization. Blood 104(7):1970–1978
Lee K-H, Wang E, Nielsen M-B, Wunderlich J, Migueles S, Connors M et al (1999) Increased vaccine-specific T cell frequency after peptide-based vaccination correlates with increased susceptibility to in vitro stimulation but does not lead to tumor regression. J Immunol 163:6292–6300
Marincola FM, Wang E, Herlyn M, Seliger B, Ferrone S (2003) Tumors as elusive targets of T cell-based active immunotherapy. Trends Immunol 24(6):335–342
Monsurro’ V, Wang E, Panelli MC, Nagorsen D, Jin P, Smith K et al (2003) Active-specific immunization against melanoma: is the problem at the receiving end? Sem Cancer Biol 13:473–480
Lee PP, Yee C, Savage PA, Fong L, Brockstedt D, Weber JS et al (1999) Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat Med 5(6):677–685
Critchley-Thorne RJ, Yan N, Nacu S, Weber J, Holmes SP, Lee PP (2007) Down-regulation of the interferon signaling pathway in T lymphocytes from patients with metastatic melanoma. PLoS Med 4(5):e176
Critchley-Thorne RJ, Simons D, Yan N, Miyahira A, Dirbas F, Johnson D et al (2009) Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci USA 106(22):9010–9015
Mortarini R, Vegetti C, Molla A, Arienti F, Ravagnani F, Maurichi A et al (2009) Impaired STAT phosphorylation in T cells from melanoma patients in response to IL-2: association with clinical stage. Clin Cancer Res 15(12):4085–4094
Marincola FM, Rivoltini L, Salgaller ML, Player M, Rosenberg SA (1996) Differential anti-MART-1/MelanA CTL activity in peripheral blood of HLA-A2 melanoma patients in comparison to healthy donors: evidence for in vivo priming by tumor cells. J Immunother 19(4):266–277
D’Souza S, Rimoldi D, Lienard D, Lejeune F, Cerottini JC, Romero P (1998) Circulating Melan-A/Mart-1 specific cytolytic T lymphocyte precursors in HLA-A2+ melanoma patients have a memory phenotype. Int J Cancer 78(6):699–706
Cormier JN, Salgaller ML, Prevette T, Rosenberg SA, Marincola FM (1996) Assessment of tumor specific immune reactivity in patients receiving immunization with the MART-1 melanoma antigen immunodominant peptide. Proc Am Ass Cancer Res 37:476
Salgaller ML, Marincola FM, Cormier JN, Rosenberg SA (1996) Immunization against epitopes in the human melanoma antigen gp100 following patient immunization with synthetic peptides. Cancer Res 56:4749–4757
Kaech SM, Hemby S, Kersh E, Ahmed R (2002) Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111:837–851
Burnet FM (1970) The concept of immunological surveillance. Prog Exp Tumor Res 13:1–27
Marincola FM, Jaffe EM, Hicklin DJ, Ferrone S (2000) Escape of human solid tumors from T cell recognition: molecular mechanisms and functional significance. Adv Immunol 74:181–273
Mantovani A, Romero P, Palucka AK, Marincola FM (2008) Tumor immunity: effector response to tumor and the influence of the microenvironment. Lancet 371(9614):771–783
Grulich AE, van Leeuwen MT, Falster MO, Vajdic CM (2007) Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet 370(9581):59–67
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C et al (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313(5795):1960–1964
Ascierto ML, De Giorgi V, Liu Q, Bedognetti D, Murtas D, Chouchane L et al (2011) An immunologic portrait of cancer. J Transl Med 9:146
Garrido F, Cabrera T, Aptsiauri N (2010) “Hard” and “soft” lesions underlying the HLA class I alterations in cancer cells: implications for immunotherapy. Int J Cancer 127(2):249–256
Seliger B (2012) Novel insights into the molecular mechanisms of HLA class I abnormalities. Cancer Immunol Immunother 61(2):249–254
Poschke I, Mougiakakos D, Kiessling R (2011) Camouflage and sabotage: tumor escape from the immune system. Cancer Immunol Immunother 60(8):1161–1171
Dunn GP, Old LJ, Schreiber RD (2004) The three Es of cancer immunoediting. Annu Rev Immunol 22:329–360
Wang E (2010) From the “delayed allergy reaction” to the “immunologic constant of rejection”. In: Wang E, Marincola FM (eds) Immunologic signatures of rejection, 1st edn. Springer, New York, pp 3–8
Marincola FM (2007) In support of descriptive studies: relevance to translational research. J Transl Med 5:21
Wang E, Marincola FM (2008) Bottom up: a modular view of immunology. Immunity 29:9–11
Wang E, Selleri S, Sabatino M, Monaco A, Pos Z, Stroncek DF et al (2008) Spontaneous and tumor-induced cancer rejection in humans. Exp Opin Biol Ther 8(3):337–349
Wang E, Miller LD, Ohnmacht GA, Mocellin S, Petersen D, Zhao Y et al (2002) Prospective molecular profiling of subcutaneous melanoma metastases suggests classifiers of immune responsiveness. Cancer Res 62:3581–3586
Panelli MC, Stashower M, Slade HB, Smith K, Norwood C, Abati A et al (2006) Sequential gene profiling of basal cell carcinomas treated with Imiquimod in a placebo-controlled study defines the requirements for tissue rejection. Genome Biol 8(1):R8
Weiss G, Grosh WW, Chianese-Bullock KA, Zhao Y, Liu H, Slingluff CL et al (2011) Molecular insights on the peripheral and intra-tumoral effects of systemic high dose rIL-2 (Aldesleukin) administration for the treatment of metastatic melanoma. Clin Cancer Res 17(23):7440–7450
Carretero R, Wang E, Rodriguez AI, Reinboth J, Ascierto ML, Engle AM et al (2011) Regression of melanoma metastases after immunotherapy is associated with activation of antigen presentation and interferon-mediated rejection genes. Int J Cancer. (Epub ahead of print)
Mocellin S, Panelli MC, Wang E, Nagorsen D, Marincola FM (2002) The dual role of IL-10. Trends Immunol 24(1):36–43
Wang E, Marincola FM (2000) A natural history of melanoma: serial gene expression analysis. Immunol Today 21(12):619–623
Cormier JN, Hijazi YM, Abati A, Fetsch P, Bettinotti M, Steinberg SM et al (1998) Heterogeneous expression of melanoma-associated antigens (MAA) and HLA-A2 in metastatic melanoma in vivo. Int J Cancer 75:517–524
Pockaj BA, Sherry RM, Wei JP, Yannelli JR, Carter CS, Leitman SF et al (1994) Localization of 111indium-labeled tumor infiltrating lymphocytes to tumor in patients receiving adoptive immunotherapy. Augmentation with cyclophosphamide and correlation with response. Cancer 73:1731–1737
Mocellin S, Ohnmacht GA, Wang E, Marincola FM (2001) Kinetics of cytokine expression in melanoma metastases classifies immune responsiveness. Int J Cancer 93:236–242
Ohnmacht GA, Wang E, Mocellin S, Abati A, Filie A, Fetsch PA et al (2001) Short term kinetics of tumor antigen expression in response to vaccination. J Immunol 167:1809–1820
Wang E, Worschech A, Marincola FM (2008) The immunologic constant of rejection. Trends Immunol 29(6):256–262
Boon T, Coulie PG, van den Eynde BJ, Van Der BP (2006) Human T cell responses against melanoma. Annu Rev Immunol 24:175–208
Lander ES (1996) The new genomics: global view of biology. Science 274:536–539
Wang E, Marincola FM (2001) cDNA microarrays and the enigma of melanoma immune responsiveness. Cancer J Sci Am 7(1):16–23
Wang E, Panelli MC, Monsurro’ V, Marincola FM (2004) Gene expression profiling of anti-cancer immune responses. Curr Opin Mol Ther 6(3):288–295
Wang E, Uccellini L, Marincola FM (2012) A genetic inference on cancer immune responsiveness. Oncoimmunology. In press
Gajewski TF (2010) Transcriptional profiling of melanoma as a potential predictive biomarker for response to immunotherapy. In Wang E, Marincola FM (eds) Signatures of rejection, 1st edn. Springer, New York, pp 229–238
Gajewski TF, Zha Y, Thurner B, Schuler G (2009) Association of gene expression profile in melanoma and survival to a dentritic cell-based vaccine. J Clin Oncol 27:A9002
Louahed J, Grusell O, Gaulis S, Coche T, Eggermont AM, Kruit W et al (2008) Expression of defined genes indentifed by pre-treatment tumor profiling: association with clinical response to GSK MAGE A-3 immunetherapeutic in metastatic melanoma patients. J Clin Oncol 26:A9045
Panelli MC, Wang E, Phan G, Puhlman M, Miller L, Ohnmacht GA et al (2002) Gene-expression profiling of the response of peripheral blood mononuclear cells and melanoma metastases to systemic IL-2 administration. Genome Biol 3(7):RESEARCH0035
Phan GQ, Attia P, Steinberg SM, White DE, Rosenberg SA (2001) Factors associated with response to high-dose interleukin-2 in patients with metastatic melanoma. J Clin Oncol 19(15):3477–3482
Sarwal M, Chua MS, Kambham N, Hsieh SC, Satterwhite T, Masek M et al (2003) Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med 349(2):125–138
Salk J (1969) Immunological paradoxes: theoretical considerations in the rejection or retention of grafts, tumors, and normal tissue. Ann N Y Acad Sci 164(2):365–380
Spivey TL, Uccellini L, Ascierto ML, Zoppoli G, De Giorgi V, Delogu LG et al (2011) Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J Transl Med 9:174
Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R et al (2005) Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 353(25):2654–2666
Gajewski TF, Louahed J, Brichard VG (2010) Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J 16(4):399–403
Sabatino M, Zhao Y, Voiculescu S, Monaco A, Robbins PF, Nickoloff BJ et al (2008) Conservation of a core of genetic alterations over a decade of recurrent melanoma supports the melanoma stem cell hypothesis. Cancer Res 68(1):222–231
Wang E, Voiculescu S, Le Poole IC, el Gamil M, Li X, Sabatino M et al (2006) Clonal persistence and evolution during a decade of recurrent melanoma. J Invest Dermatol 126(6):1372–1377
Marincola FM (2011) The trouble with translational medicine. J Int Med 270(2):123–127
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study is based on a presentation given at the Ninth Annual Meeting of the Association for Cancer Immunotherapy (CIMT), held in Mainz, Germany, 25–27th May, 2011. It follows a CII series of Focussed Research Reviews and meeting report published in Cancer Immunology, Immunotherapy, Volume 61, Number 1, in January 2012.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, E., Tomei, S. & Marincola, F.M. Reflections upon human cancer immune responsiveness to T cell-based therapy. Cancer Immunol Immunother 61, 761–770 (2012). https://doi.org/10.1007/s00262-012-1274-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-012-1274-9