Abstract
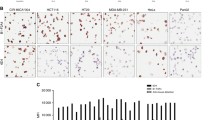
Immunoscreening of an Ewing’s family of tumour (EFT)-derived cDNA library using formerly described EFT-specific antibodies led to the isolation of a 3.5 kb cDNA, named Ewing’s tumour-associated antigen 16 (ETAA16). The ETAA16 cDNA shows no homology to any functionally characterised human gene. Only a bovine cDNA expressed in bovine testis and hepatocytes is functionally characterised as it encodes for a junction plaque associated protein and showed a homology of 69.9% at amino acid level to ETAA16. The human cDNA encodes for a 926 amino acid tumour antigen with a calculated molecular weight of 103 kDa. The epitope of the ETAA16-specific antibody, Ak16, covers the central region of the protein which is part of an extra cellular domain. The human ETAA16 gene locus has been assigned to chromosome 2p13-15 by FISH analyses and is confirmed by the human genome sequencing project. As demonstrated by flow cytometry, the cell surface expression of ETAA16 antigen is restricted to ET cell lines and not expressed on other small blue round cell tumours or other kind of tumour. RT-PCR analysis revealed a high expression of ETAA16 in brain, liver and kidney while lung and heart were negative. Immunohistochemistry showed an intracellular expression of ETAA16 in different kind of non-Ewing tumour tissues. These results suggest that ETAA16 may function as a tumour-specific cell surface antigen in EFTs.







Similar content being viewed by others
Abbreviations
- DAB:
-
Diaminobenzidine
- EFT:
-
Ewing’s family of tumours
- ET:
-
Ewing’s tumour
- ETAA16:
-
Ewing’s tumour-associated antigen 16
- ETS:
-
E26 transformation specific
- EST:
-
Expressed sequence tag
- EWS:
-
Ewing’s sarcoma
- FISH:
-
Fluorescence in situ hybridisation
- FLI-1:
-
Friend leukemia integration site-1
- MAB:
-
Monoclonal antibody
- MFI:
-
Mean fluorescence intensity
- ORF:
-
Open reading frame
- PALS:
-
Periarteriolar lymphoid sheath
- PNET:
-
Primitive neuroectodermal tumour
- UTR:
-
Untranslated region
References
Ambros IM, Ambros PF, Strehl S, Kovar H, Gadner H, Salzer-Kuntschik M (1991) MIC2 is a specific marker for Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing’s sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer 67:1886
Arvand A, Bastians H, Welford SM, Thompson AD, Ruderman JV, Denny CT (1998) EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene 17:2039
Arvand A, Denny CT (2001) Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene 20:5747
Borowski A, van Valen F, et al. (1999) Monomorphic HLA class I-(non-A, non-B) expression on Ewing’s tumor cell lines, modulation by TNF-α and IFN-γ. Immunobiology 200:1
Braun BS, Frieden R, Lessnick SL, May WA, Denny CT (1995) Identification of target genes for the Ewing’s sarcoma EWS/FLI fusion protein by representational difference analysis. Mol Cell Biol 15:4623
Cavazzana AO, Miser JS, Jefferson J, Triche TJ (1987) Experimental evidence for a neural origin of Ewing’s sarcoma of bone. Am J Pathol 127:507
Chomzynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156
Dixkens C, Posseckert G, Keller T, Hameister H (1998) Structural analysis of the amplified IFN-beta and DHFR genes in a Chinese hamster ovary cell line using multicolour FISH analysis. Chromosome Res 6:329
Franchi A, Pasquinelli G, et al. (2001) Immunohistochemical and ultrastructural investigation of neural differentiation in Ewing sarcoma/PNET of bone and soft tissues. Ultrastruct Pathol 25:219
Granowetter L (1995) Ewing’s sarcoma and extracranial peripheral neuroectodermal tumors. Curr Opin Oncol 7:355
Hahm KB, Cho K, et al. (1999) Repression of the gene encoding the TGF-beta type II receptor is a major target of the EWS-FLI1 oncoprotein. Nat Genet 23:222
Jankowski J, Holst MI, Liebig C, Oberdick J, Baader SL (2004) Engrailed-2 negatively regulates the onset of perinatal Purkinje cell differentiation. J Comp Neurol 472:87
Jürgens HF (1994) Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. Curr Opin Oncol 6:391
Kato K, Arai K, et al. (2000) Epithelioid leiomyosarcoma in a non-immunocompromised infant: additional differential diagnosis of pediatric “Round Cell Tumors”. Mod Pathol 13:1156
Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283
Kozak M (1996) Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome 7:563
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105
Lander ES, Linton LM et al. (2001) Initial sequencing and analysis of the human genome. Nature 409:860
Laws HJ, Burdach S, et al. (1999) Multimodality diagnostics and megatherapy in poor prognosis Ewing’s tumor patients. a single-center report. Strahlenther Onkol 175:488
May WA, Arvand A, Thompson AD, Braun BS, Wright M, Denny CT (1997) EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat Genet 17:495
Melief CJ, Toes RE, Medema JP, van der Burg SH, Ossendorp F, Offringa R (2000) Strategies for immunotherapy of cancer. Adv Immunol 75:235
Pagani A, Fischer-Colbrie R, Eder U, Pellin A, Llombart-Bosch A, Bussolati G (1995) Neural and mesenchymal differentiations in Ewing’s sarcoma cell lines. Morphological, immunophenotypic, molecular biological and cytogenetic evidence. Int J Cancer 63:738
Paulussen M, Ahrens S, et al. (2001) Localized Ewing tumor of bone:final results of the cooperative Ewing’s Sarcoma Study CESS 86. J Clin Oncol 19:1818
Remy P, Baltzinger M (2000) The Ets-transcription factor family in embryonic development: lessons from the amphibian and bird. Oncogene 19:6417
Ribas A, Butterfield LH, Glaspy JA, Economou JS (2003) Current developments in cancer vaccines and cellular immunotherapy. J Clin Oncol 21:2415
Sambrook J, Maniatis T, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbour Laboratory Press, New York
Schaefer KL, Wai DH, et al. (2002) Characterization of the malignant melanoma of soft-parts cell line GG-62 by expression analysis using DNA microarrays. Virchows Arch 440:476
Shi LR, Eichelbauer D, Borchard F, Jurgens H, Gobel U, Schneider EM (1994) Specificity and function of monoclonal antibodies directed against Ewing sarcoma cells. Cancer Immunol Immunother 38:208
Slaper-Cortenbach IC, Admiraal LG, Kerr JM, van Leeuwen EF, von dem Borne AE, Tetteroo PA (1988) Flow-cytometric detection of terminal deoxynucleotidyl transferase and other intracellular antigens in combination with membrane antigens in acute lymphatic leukemias. Blood 72:1639
Spieker N, van Sluis P, et al. (2001) The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics 71:214
Staege MS, Hutter C, et al. (2004) DNA Microarrays reveal relationship of ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res 64:8213
Stratakis CA, Taymans SE (1998) Structure of the gene coding for calcineurin B (PPP3R1) and mapping to D2S358-D2S1778 (chromosomal region 2p15). DNA Seq 9:227
Urban JL, Schreiber H (1992) Tumor Antigens. Annu Rev Immunol 10:617
van Valen F (1999) Ewing’s sarcoma family of tumors. In: Masters JRW, Palsson B (eds), Human cell culture, vol. 1. Kluwer, Dordrecht pp 55–85
Vlaeminck-Guillem V, Carrere S, Dewitte F, Stehelin D, Desbiens X, Duterque-Coquillaud M (2000) The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech Dev 91:331
Watanabe G, Nishimori H, et al. (2003) Induction of tenascin-C by tumor-specific EWS-ETS fusion genes. Genes Chromosomes Cancer 36:224
Zwerner JP, May WA (2001) PDGF-C is an EWS/FLI induced transforming growth factor in Ewing family tumors. Oncogene 20:626
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft DI 747/3–1; Di747/3–2 and the Krebshilfe, Dr. Mildred Scheel Stiftung: 10–1859/Di, W26/94/Schn2 and the Elterninitiative Kinderkrebsklinik e.V. The authors thank Professor Horst Hameister, Department of Human Genetics, University of Ulm for performing the FISH analyses and his contribution in data analysis; stud. Biol. Stefanie Borkens Heinrich Heine University Düsseldorf for excellent technical assistance, Prof. Guido Reifenberger and Prof. Christopher Poremba, Institut for Pathology University Medical Center Düsseldorf for providing various cryopreserved tumour tissues. The authors also wish to thank Bettina Stahl (Anaesthesiology, Ulm) and Whilhelma Langmann (Department of Anatomy, Bonn) for their help with the immunohistochemical staining and also Dr. Kay Hofmann, Memorec, Cologne, Germany, for the bioinformatics analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Borowski, A., Dirksen, U., Lixin, L. et al. Structure and function of ETAA16: a novel cell surface antigen in Ewing’s tumours. Cancer Immunol Immunother 55, 363–374 (2006). https://doi.org/10.1007/s00262-005-0017-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-005-0017-6