Abstract
Purpose
To achieve prenatal prediction of placenta accreta spectrum (PAS) by combining clinical model, radiomics model, and deep learning model using T2-weighted images (T2WI), and to objectively evaluate the performance of the prediction through multicenter validation.
Methods
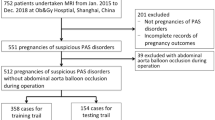
A total of 407 pregnant women from two centers undergoing preoperative magnetic resonance imaging (MRI) were retrospectively recruited. The patients from institution I were divided into a training cohort (n = 298) and a validation cohort (n = 75), while patients from institution II served as the external test cohort (n = 34). In this study, we built a clinical prediction model using patient clinical data, a radiomics model based on selected key features, and a deep learning model by mining deep semantic features. Based on this, we developed a combined model by ensembling the prediction results of the three models mentioned above to achieve prenatal prediction of PAS. The performance of these predictive models was evaluated with respect to discrimination, calibration, and clinical usefulness.
Results
The combined model achieved AUCs of 0.872 (95% confidence interval, 0.843 to 0.908) in the validation cohort and 0.857 (0.808 to 0.894) in the external test cohort, both of which outperformed the other models. The calibration curves demonstrated excellent consistency in the validation cohort and the external test cohort, and the decision curves indicated high clinical usefulness.
Conclusion
By using preoperative clinical information and MRI images, the combined model can accurately predict PAS by ensembling clinical model, radiomics model, and deep learning model.
Graphical abstract









Similar content being viewed by others
Abbreviations
- PAS:
-
Placenta accreta spectrum
- T2WI:
-
T2-weighted images
- MRI:
-
Magnetic resonance imaging
- CI:
-
Confidence interval
- ROI:
-
Regions of interest
- SVM:
-
Support vector machine
- LASSO:
-
Least absolute shrinkage and selection operator
- ROC:
-
Receiver operating characteristics
- AUC:
-
Area under curve
References
Shamshirsaz AA, Fox KA, Erfani H, et al (2017) Multidisciplinary team learning in the management of the morbidly adherent placenta: outcome improvements overtime. Am Journal Obstet Gynecol 216: 612.e1-612.e5. https://doi.org/10.1016/j.ajog.2017.02.016
Solomon CG, Silver RM, Branch DW (2018) Placenta accreta spectrum. New Engl J Med 378: 1529-1536. https://doi.org/10.1056/NEJMcp1709324
Ueno Y, Kitajima K, Kawakami F, et al (2014) Novel MRI finding for diagnosis of invasive placenta praevia: evaluation of findings for 65 patients using clinical and histopathological correlations. Eur Radiol 24: 881-888. https://doi.org/10.1007/s00330-013-3076-7
Zeng C, Yang M, Ding Y, et al (2018). Placenta accreta spectrum disorder trends in the context of the universal two‐child policy in China and the risk of hysterectomy. Int J Gynecol Obstet 140: 312-318. https://doi.org/10.1002/ijgo.12418
Jauniaux E, Chantraine F, Silver RM, et al (2018) FIGO consensus guidelines on placenta accreta spectrum disorders: Epidemiology. Int J Gynecol Obstet 140: 265-273. https://doi.org/10.1002/ijgo.12407
Kapoor H, Hanaoka M, Dawkins A, et al (2021) Review of MRI imaging for placenta accreta spectrum: pathophysiologic insights, imaging signs, and recent developments. Placenta 104: 31-39. https://doi.org/10.1016/j.placenta.2020.11.004
Jauniaux E, Collins S, Burton GJ (2017) Placenta accreta spectrum: pathophysiology and evidence-based anatomy for prenatal ultrasound imaging. Am J Obstet Gyneecol 218: 75-87. https://doi.org/10.1016/j.ajog.2017.05.067
Do QN, Lewis MA, Yin X, et al (2020) MRI of the Placenta Accreta Spectrum (PAS) Disorder: Radiomics Analysis Correlates With Surgical and Pathological Outcome. J Magn Reson Imging 51: 936-946. https://doi.org/10.1002/jmri.26883
Pizzi AD, Tavoletta A, Narciso R, et al (2019) Prenatal planning of placenta previa: diagnostic accuracy of a novel MRI-based prediction model for placenta accreta spectrum (PAS) and clinical outcome. Abdominal Radiology 44:1873-1882. https://doi.org/10.1007/s00261-018-1882-8
Chen T, Xu X Q, Shi H B, et al (2017) Conventional MRI features for predicting the clinical outcome of patients with invasive placenta. Diagn Interv Radiol 23: 173. https://doi.org/10.5152/dir.2016.16412
Ueno Y, Maeda T, Tanaka U, et al (2016) Evaluation of interobserver variability and diagnostic performance of developed MRI‐based radiological scoring system for invasive placenta previa. J Magn Reson Imaging 44: 573-583. https://doi.org/10.1002/jmri.25184
Siauve N (2019) How and why should the radiologist look at the placenta? Eur Radiol 29: 6149-6151. https://doi.org/10.1007/s00330-019-06373-8
Wu Q, Yao K, Liu Z, et al (2019) Radiomics analysis of placenta on T2WI facilitates prediction of postpartum haemorrhage: a multicentre study. EBioMedicine, 50: 355-365. https://doi.org/10.1016/j.ebiom.2019.11.010
Romeo V, Ricciardi C, Cuocolo R, et al (2019) Machine learning analysis of MRI-derived texture features to predict placenta accreta spectrum in patients with placenta previa. Magn Resone Imaging 64: 71-76. https://doi.org/10.1016/j.mri.2019.05.017
Sun H, Qu H, Chen L, et al (2019) Identification of suspicious invasive placentation based on clinical MRI data using textural features and automated machine learning. Eur Radiol 29: 6152-6162. https://doi.org/10.1007/s00330-019-06372-9
Ren H, Mori N, Mugikura S, et al (2021) Prediction of placenta accreta spectrum using texture analysis on coronal and sagittal T2-weighted imaging. Abdominal Radiology 46: 5344-5352. https://doi.org/10.1007/s00261-021-03226-1
Huang B, Tian J, Zhang H, et al (2020) Deep semantic segmentation feature-based radiomics for the classification tasks in medical image analysis. IEEE Journal of Biomedical and Health Informatics 25: 2655-2664. https://doi.org/10.1109/JBHI.2020.3043236
Wei J, Cheng J, Gu D, et al (2021) Deep learning‐based radiomics predicts response to chemotherapy in colorectal liver metastases. Med Phys 48: 513-522. https://doi.org/10.1002/mp.14563
Song J, Ding C, Huang Q, et al (2021) Deep learning predicts epidermal growth factor receptor mutation subtypes in lung adenocarcinoma. Med Phys 48: 7891-7899. https://doi.org/10.1002/mp.15307
Xuan R, Li T, Wang Y, et al (2021) Prenatal prediction and typing of placental invasion using MRI deep and radiomic features. BioMedical Engineering OnLine 20: 1-18. https://doi.org/10.1186/s12938-021-00893-5
Shao Q, Xuan R, Wang Y, et al (2021) Deep learning and radiomics analysis for prediction of placenta invasion based on T2WI. Math Biosci Eng 18: 6198-6215. https://doi.org/10.3934/mbe.2021310
Li C, Qin Y, Zhang W, et al (2022) Deep learning‐based AI model for signet‐ring cell carcinoma diagnosis and chemotherapy response prediction in gastric cancer. Med phys 49: 1535-1546. https://doi.org/10.1002/mp.15437
Wang S, Dong D, Li L, et al (2021) A deep learning radiomics model to identify poor outcome in COVID-19 patients with underlying health conditions: a multicenter study. IEEE Journal of Biomedical and Health Informatics 25: 2353-2362. https://doi.org/10.1109/JBHI.2021.3076086
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, pp. 234-241
He K, Zhang X, Ren S, et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 770–778
Perkinsm NJ, Schisterman EF (2006) The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. American journal of epidemiology 163: 670-675. https://doi.org/10.1093/aje/kwj292
Imafuku H, Tanimura K, Shi Y, et al (2021) Clinical factors associated with a placenta accreta spectrum. Placenta 112: 180-184. https://doi.org/10.1016/j.placenta.2021.08.001
Funding
This work was supported in part by the Natural Science Foundation of Zhejiang Province under Grant no. LY20H180003, the Public Welfare Science and Technology Project of Ningbo under Grant no. 2022S043.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ye, Z., Xuan, R., Ouyang, M. et al. Prediction of placenta accreta spectrum by combining deep learning and radiomics using T2WI: a multicenter study. Abdom Radiol 47, 4205–4218 (2022). https://doi.org/10.1007/s00261-022-03673-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-022-03673-4