Abstract
Objective
The aim of this study was to investigate whether intraplacental texture features from routine placental MRI can objectively and accurately predict invasive placentation.
Material and methods
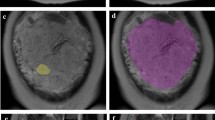
This retrospective study includes 99 pregnant women with pathologically confirmed placental invasion and 56 pregnant women with simple placenta previa. All participants underwent magnetic resonance imaging after 24 gestational weeks. The placenta was segmented in sagittal images from both turbo spin echo (TSE) and balanced turbo field echo (bTFE) sequences. Textural features were extracted from the both original and Laplacian of Gaussian (LoG)-filtered MRI images. An automated machine learning algorithm was applied to the extracted feature sets to obtain the optimal preprocessing steps, classification algorithm, and corresponding hyper-parameters.
Results
A gradient boosting classifier using all textual features from original and LoG-filtered TSE images and bTFE images identified by the automated machine learning algorithm achieved the optimal performance with sensitivity, specificity, accuracy, and area under ROC curve (AUC) of 100%, 88.5%, 95.2%, and 0.98 in the prediction of placental invasion. In addition, textural features that contributed to the prediction of placental invasion differ from the features significantly affected by normal placenta maturation.
Conclusions
Quantifying intraplacental heterogeneity using LoG filtration and texture analysis highlights the different heterogeneous appearance caused by abnormal placentation relative to normal maturation. The predictive model derived from automated machine learning yielded good performance, indicating the proposed radiomic analysis pipeline can accurately predict placental invasion and facilitate clinical decision-making for pregnant women with suspicious placental invasion.
Key Points
• The intraplacental texture features have high efficiency in prediction of invasive placentation after 24 gestational weeks.
• The features with dominated predictive power did not overlap with the features significantly affected by gestational age.





Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the ROC curve
- bTFE:
-
Balanced turbo field echo
- LoG:
-
Laplacian of Gaussian
- PI:
-
Placental invasion
- SPP:
-
Simple placenta previa
- TSE:
-
Turbo spin echo
References
Oyelese Y, Smulian JC (2006) Placenta previa, placenta accreta, and vasa previa. Obstet Gynecol 107:927–941. https://doi.org/10.1097/01.AOG.0000207559.15715.98
Garmi G, Salim R (2012) Epidemiology, etiology, diagnosis, and management of placenta accreta. Obstet Gynecol Int 2012:1–7. https://doi.org/10.1155/2012/873929
Baughman WC, Corteville JE, Shah RR (2008) Placenta accreta: spectrum of US and MR imaging findings. Radiographics 28:1905–1916. https://doi.org/10.1148/rg.287085060
Kocher MR, Sheafor DH, Bruner E et al (2017) Diagnosis of abnormally invasive posterior placentation: the role of MR imaging. Radiol Case Rep 12:295–299. https://doi.org/10.1016/j.radcr.2017.01.014
Delli Pizzi A, Tavoletta A, Narciso R et al (2019) Prenatal planning of placenta previa: diagnostic accuracy of a novel MRI-based prediction model for placenta accreta spectrum (PAS) and clinical outcome. Abdom Radiol 44:1873–1882. https://doi.org/10.1007/s00261-018-1882-8
D’Antonio F, Iacovella C, Palacios-Jaraquemada J et al (2014) Prenatal identification of invasive placentation using magnetic resonance imaging: systematic review and meta-analysis. Ultrasound Obstet Gynecol 44:8–16. https://doi.org/10.1002/uog.13327
Alamo L, Anaye A, Rey J et al (2013) Detection of suspected placental invasion by MRI: do the results depend on observer’ experience? Eur J Radiol 82:e51–e57. https://doi.org/10.1016/j.ejrad.2012.08.022
Ueno Y, Kitajima K, Kawakami F et al (2014) Novel MRI finding for diagnosis of invasive placenta praevia: evaluation of findings for 65 patients using clinical and histopathological correlations. Eur Radiol 24:881–888. https://doi.org/10.1007/s00330-013-3076-7
Lax A, Prince MR, Mennitt KW et al (2007) The value of specific MRI features in the evaluation of suspected placental invasion. Magn Reson Imaging 25:87–93. https://doi.org/10.1016/j.mri.2006.10.007
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Langs G, Röhrich S, Hofmanninger J et al (2018) Machine learning: from radiomics to discovery and routine. Radiologe 58:1–6. https://doi.org/10.1007/s00117-018-0407-3
Castellano G, Bonilha L, Li LM, Cendes F (2004) Texture analysis of medical images. Clin Radiol 59:1061–1069. https://doi.org/10.1016/j.crad.2004.07.008
Horowitz JM, Berggruen S, McCarthy RJ et al (2015) When timing is everything: are placental MRI examinations performed before 24 weeks’ gestational age reliable? Am J Roentgenol 205:685–692. https://doi.org/10.2214/AJR.14.14134
Wang G, Zuluaga MA, Pratt R et al (2016) Slic-Seg: a minimally interactive segmentation of the placenta from sparse and motion-corrupted fetal MRI in multiple views. Med Image Anal 34:137–147. https://doi.org/10.1016/j.media.2016.04.009
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128. https://doi.org/10.1016/j.neuroimage.2006.01.015
Van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Yip SSF, Aerts HJWL (2016) Applications and limitations of radiomics. Phys Med Biol 61:R150–R166. https://doi.org/10.1088/0031-9155/61/13/R150
Fadl S, Moshiri M, Fligner CL et al (2017) Placental imaging: normal appearance with review of pathologic findings. Radiographics 37:979–998. https://doi.org/10.1148/rg.2017160155
Dukart J, Schroeter ML, Mueller K (2011) Age correction in dementia – matching to a healthy brain. PLoS One 6:e22193. https://doi.org/10.1371/journal.pone.0022193
Olson RS, Bartley N, Urbanowicz RJ, Moore JH (2016) Evaluation of a Tree-based Pipeline Optimization Tool for automating data science. Proc 2016 Genet Evol Comput Conf - GECCO ‘16 485–492. https://doi.org/10.1145/2908812.2908918
Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20:37–46. https://doi.org/10.1177/001316446002000104
Natekin A, Knoll A (2013) Gradient boosting machines, a tutorial. Front Neurorobot 7. https://doi.org/10.3389/fnbot.2013.00021
Wang S, Summers RM (2012) Machine learning and radiology. Med Image Anal 16:933–951. https://doi.org/10.1016/j.media.2012.02.005
Parmar C, Grossmann P, Bussink J et al (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087. https://doi.org/10.1038/srep13087
Larue RTHMHM, Defraene G, De Ruysscher D et al (2017) Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 90:20160665. https://doi.org/10.1259/bjr.20160665
Scheffler K, Lehnhardt S (2003) Principles and applications of balanced SSFP techniques. Eur Radiol 13:2409–2418. https://doi.org/10.1007/s00330-003-1957-x
Neycenssac F (1993) Contrast enhancement using the Laplacian-of-a-Gaussian filter. CVGIP Graph Model Image Process 55:447–463. https://doi.org/10.1006/cgip.1993.1034
Rahaim NSA, Whitby EH (2015) The MRI features of placental adhesion disorder and their diagnostic significance: systematic review. Clin Radiol 70:917–925. https://doi.org/10.1016/j.crad.2015.04.010
Tang X (1998) Texture information in run-length matrices. IEEE Trans Image Process 7:1602–1609. https://doi.org/10.1109/83.725367
Blaicher W, Brugger PC, Mittermayer C et al (2006) Magnetic resonance imaging of the normal placenta. Eur J Radiol 57:256–260. https://doi.org/10.1016/j.ejrad.2005.11.025
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3:610–621. https://doi.org/10.1109/TSMC.1973.4309314
Funding
This study was supported by the National Natural Science Foundation of China (81601458), the Ministry of Science and Technology of China (2016YFC0100803, 2018YFC1004603), the Health and Family Planning Commission of Sichuan Province (16ZD017), and Bureau of Science and Technology of Chengdu City (2014-HM01-00314-SF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is S. Zhou.
Conflict of interest
The authors declare that they have no conflict of interest.
Statistics and biometry
One of the authors (H. Sun) has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• cross-sectional study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 31312 kb)
Rights and permissions
About this article
Cite this article
Sun, H., Qu, H., Chen, L. et al. Identification of suspicious invasive placentation based on clinical MRI data using textural features and automated machine learning. Eur Radiol 29, 6152–6162 (2019). https://doi.org/10.1007/s00330-019-06372-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06372-9