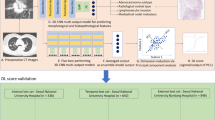
Abstract
Purpose
No consensus on a grading system for invasive lung adenocarcinoma had been built over a long period of time. Until October 2020, a novel grading system was proposed to quantify the whole landscape of histologic subtypes and proportions of pulmonary adenocarcinomas. This study aims to develop a deep learning grading signature (DLGS) based on positron emission tomography/computed tomography (PET/CT) to personalize surgical treatments for clinical stage I invasive lung adenocarcinoma and explore the biologic basis under its prediction.
Methods
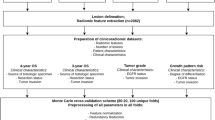
A total of 2638 patients with clinical stage I invasive lung adenocarcinoma from 4 medical centers were retrospectively included to construct and validate the DLGS. The predictive performance of the DLGS was evaluated by the area under the receiver operating characteristic curve (AUC), its potential to optimize surgical treatments was investigated via survival analyses in risk groups defined by the DLGS, and its biological basis was explored by comparing histologic patterns, genotypic alternations, genetic pathways, and infiltration of immune cells in microenvironments between risk groups.
Results
The DLGS to predict grade 3 achieved AUCs of 0.862, 0.844, and 0.851 in the validation set (n = 497), external cohort (n = 382), and prospective cohort (n = 600), respectively, which were significantly better than 0.814, 0.810, and 0.806 of the PET model, 0.813, 0.795, and 0.824 of the CT model, and 0.762, 0.734, and 0.751 of the clinical model. Additionally, for DLGS-defined high-risk population, lobectomy yielded an improved prognosis compared to sublobectomy p = 0.085 for overall survival [OS] and p = 0.038 for recurrence-free survival [RFS]) and systematic nodal dissection conferred a superior prognosis to limited nodal dissection (p = 0.001 for OS and p = 0.041 for RFS).
Conclusion
The DLGS harbors the potential to predict the histologic grade and personalize the surgical treatments for clinical stage I invasive lung adenocarcinoma. Its applicability to other territories should be further validated by a larger international study.





Similar content being viewed by others
Data availability
The imaging data and clinical information in the current study are not publicly available for patient privacy purposes but are available from the corresponding authors upon reasonable request. The proposed source code would be provided at GitHub.
References
Helpap B, Ringli D, Tonhauser J, Poser I, Breul J, Gevensleben H, et al. The significance of accurate determination of gleason score for therapeutic options and prognosis of prostate cancer. Pathol Oncol Res. 2016;22:349–56. https://doi.org/10.1007/s12253-015-0013-x.
Rabe K, Snir OL, Bossuyt V, Harigopal M, Celli R, Reisenbichler ES. Interobserver variability in breast carcinoma grading results in prognostic stage differences. Hum Pathol. 2019;94:51–7. https://doi.org/10.1016/j.humpath.2019.09.006.
Rice-Stitt T, Valencia-Guerrero A, Cornejo KM, Wu CL. Updates in histologic grading of urologic neoplasms. Arch Pathol Lab Med. 2020;144:335–43. https://doi.org/10.5858/arpa.2019-0551-RA.
Warth A, Muley T, Kossakowski C, Stenzinger A, Schirmacher P, Dienemann H, et al. Prognostic impact and clinicopathological correlations of the cribriform pattern in pulmonary adenocarcinoma. J Thorac Oncol. 2015;10:638–44. https://doi.org/10.1097/JTO.0000000000000490.
Warth A, Muley T, Meister M, Stenzinger A, Thomas M, Schirmacher P, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol. 2012;30:1438–46. https://doi.org/10.1200/jco.2011.37.2185.
Yoshizawa A, Sumiyoshi S, Sonobe M, Kobayashi M, Fujimoto M, Kawakami F, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol. 2013;8:52–61. https://doi.org/10.1097/JTO.0b013e3182769aa8.
Moreira AL, Ocampo PSS, Xia Y, Zhong H, Russell PA, Minami Y, et al. A grading system for invasive pulmonary adenocarcinoma: a proposal from the international association for the study of lung cancer pathology committee. J Thorac Oncol. 2020;15:1599–610. https://doi.org/10.1016/j.jtho.2020.06.001.
Deng C, Zheng Q, Zhang Y, Jin Y, Shen X, Nie X, et al. Validation of the novel international association for the study of lung cancer grading system for invasive pulmonary adenocarcinoma and association with common driver mutations. J Thorac Oncol. 2021;16:1684–93. https://doi.org/10.1016/j.jtho.2021.07.006.
Hou L, Wang T, Chen D, She Y, Deng J, Yang M, et al. Prognostic and predictive value of the newly proposed grading system of invasive pulmonary adenocarcinoma in Chinese patients: a retrospective multicohort study. Mod Pathol. 2022. https://doi.org/10.1038/s41379-021-00994-5.
Fujikawa R, Muraoka Y, Kashima J, Yoshida Y, Ito K, Watanabe H, et al. Clinicopathologic and genotypic features of lung adenocarcinoma characterized by the international association for the study of lung cancer grading system. J Thorac Oncol. 2022;17:700–7. https://doi.org/10.1016/j.jtho.2022.02.005.
Su H, Xie H, Dai C, Zhao S, Xie D, She Y, et al. Procedure-specific prognostic impact of micropapillary subtype may guide resection strategy in small-sized lung adenocarcinomas: a multicenter study. Ther Adv Med Oncol. 2020;12:1758835920937893. https://doi.org/10.1177/1758835920937893.
Nitadori J, Bograd AJ, Kadota K, Sima CS, Rizk NP, Morales EA, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J Natl Cancer Inst. 2013;105:1212–20. https://doi.org/10.1093/jnci/djt166.
Sun W, Su H, Liu J, Zhang L, Li M, Xie H, et al. Impact of histological components on selecting limited lymphadenectomy for lung adenocarcinoma ≤ 2 cm. Lung Cancer. 2020;150:36–43. https://doi.org/10.1016/j.lungcan.2020.09.016.
Moon Y, Kim KS, Lee KY, Sung SW, Kim YK, Park JK. Clinicopathologic factors associated with occult lymph node metastasis in patients with clinically diagnosed N0 lung adenocarcinoma. Ann Thorac Surg. 2016;101:1928–35. https://doi.org/10.1016/j.athoracsur.2015.11.056.
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60. https://doi.org/10.1097/jto.0000000000000630.
Travis WD, Rekhtman N, Riley GJ, Geisinger KR, Asamura H, Brambilla E, et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: a paradigm shift. J Thorac Oncol. 2010;5:411–4. https://doi.org/10.1097/JTO.0b013e3181d57f6e.
Cataluña JJ, Perpiñá M, Greses JV, Calvo V, Padilla JD, París F. Cell type accuracy of bronchial biopsy specimens in primary lung cancer. Chest. 1996;109:1199–203. https://doi.org/10.1378/chest.109.5.1199.
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10. https://doi.org/10.1038/s41568-018-0016-5.
Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636–42. https://doi.org/10.1001/jamaoncol.2016.2631.
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. https://doi.org/10.1038/ncomms5006.
Chen HHW, Chiu N-T, Su W-C, Guo H-R, Lee B-F. Prognostic value of whole-body total lesion glycolysis at pretreatment FDG PET/CT in non–small cell lung cancer. Radiology. 2012;264:559–66. https://doi.org/10.1148/radiol.12111148.
Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC lung cancer staging project. J Thorac Oncol. 2008;3:6–12. https://doi.org/10.1097/JTO.0b013e31815e6d6b.
Nair VS, Barnett PG, Ananth L, Gould MK. PET scan 18F-fluorodeoxyglucose uptake and prognosis in patients with resected clinical stage IA non-small cell lung cancer. Chest. 2010;137:1150–6. https://doi.org/10.1378/chest.09-2356.
Nie P, Yang G, Wang N, Yan L, Miao W, Duan Y, et al. Additional value of metabolic parameters to PET/CT-based radiomics nomogram in predicting lymphovascular invasion and outcome in lung adenocarcinoma. Eur J Nucl Med Mol Imaging. 2021;48:217–30. https://doi.org/10.1007/s00259-020-04747-5.
Hyun SH, Ahn MS, Koh YW, Lee SJ. A machine-learning approach using PET-based radiomics to predict the histological subtypes of lung cancer. Clin Nucl Med. 2019;44:956–60. https://doi.org/10.1097/rlu.0000000000002810.
Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45:207–17. https://doi.org/10.1007/s00259-017-3837-7.
Huang B, Sollee J, Luo YH, Reddy A, Zhong Z, Wu J, et al. Prediction of lung malignancy progression and survival with machine learning based on pre-treatment FDG-PET/CT. EBioMedicine. 2022;82:104127. https://doi.org/10.1016/j.ebiom.2022.104127.
Mu W, Tunali I, Gray JE, Qi J, Schabath MB, Gillies RJ. Radiomics of 18F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur J Nucl Med Mol Imaging. 2020;47:1168–82. https://doi.org/10.1007/s00259-019-04625-9.
Dissaux G, Visvikis D, Da-Ano R, Pradier O, Chajon E, Barillot I, et al. Pretreatment (18)F-FDG PET/CT radiomics predict local recurrence in patients treated with stereotactic body radiotherapy for early-stage non-small cell lung cancer: a multicentric study. J Nucl Med. 2020;61:814–20. https://doi.org/10.2967/jnumed.119.228106.
Bassi M, Russomando A, Vannucci J, Ciardiello A, Dolciami M, Ricci P, et al. Role of radiomics in predicting lung cancer spread through air spaces in a heterogeneous dataset. Transl Lung Cancer Res. 2022;11:560–71. https://doi.org/10.21037/tlcr-21-895.
Song SH, Park H, Lee G, Lee HY, Sohn I, Kim HS, et al. Imaging phenotyping using radiomics to predict micropapillary pattern within lung adenocarcinoma. J Thorac Oncol. 2017;12:624–32. https://doi.org/10.1016/j.jtho.2016.11.2230.
Park S, Lee SM, Noh HN, Hwang HJ, Kim S, Do KH, et al. Differentiation of predominant subtypes of lung adenocarcinoma using a quantitative radiomics approach on CT. Eur Radiol. 2020;30:4883–92. https://doi.org/10.1007/s00330-020-06805-w.
Kuhn E, Morbini P, Cancellieri A, Damiani S, Cavazza A, Comin CE. Adenocarcinoma classification: patterns and prognosis. Pathologica. 2018;110:5–11.
Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. https://doi.org/10.1038/nrg1427.
Nogués L, Palacios-García J, Reglero C, Rivas V, Neves M, Ribas C, et al. G protein-coupled receptor kinases (GRKs) in tumorigenesis and cancer progression: GPCR regulators and signaling hubs. Semin Cancer Biol. 2018;48:78–90. https://doi.org/10.1016/j.semcancer.2017.04.013.
Liebermann DA, Hoffman B. Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia. 2002;16:527–41. https://doi.org/10.1038/sj.leu.2402477.
Vitale I, Manic G, Coussens LM, Kroemer G, Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30:36–50. https://doi.org/10.1016/j.cmet.2019.06.001.
Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER Research Data, 8 Registries, Nov 2021 Sub (1975–2020) - Linked To County Attributes - Time Dependent (1990–2020) Income/Rurality, 1969–2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2023, based on the November 2022 submission. Accessed 1 Sep 2023
Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52:177–86. https://doi.org/10.1038/s41588-019-0569-6.
Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154–62. https://doi.org/10.1097/jto.0000000000000033.
Funding
This study was supported by National Key Research and Development Program of China (2022YFC2407401); National Natural Science Foundation of China (92259205, 82102126, 82272943); Science and Technology Commission of Shanghai Municipality(21YF1438200); Clinical Research Foundation of Shanghai Pulmonary Hospital (SKPY2021008); Investigator-Initiated Trial of Shanghai Pulmonary Hospital (2021LY1144, 2023LY0310); Ningbo Top Medical and Health Research Program (2022030208); and Medicine and Public Health Scientific Projects in Zhejiang Province (2020KY270)
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Yifan Zhong, Chuang Cai, and Tao Chen. The first draft of the manuscript was written by Yifan Zhong, Chuang Cai, and Tao Chen, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This study was conducted under the approval of the Institutional Review Board of Shanghai Pulmonary Hospital, The First Affiliated Hospital of Nanchang University, the Affiliated Hospital of Zunyi Medical College, and Ningbo HwaMei Hospital.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhong, Y., Cai, C., Chen, T. et al. PET/CT-based deep learning grading signature to optimize surgical decisions for clinical stage I invasive lung adenocarcinoma and biologic basis under its prediction: a multicenter study. Eur J Nucl Med Mol Imaging 51, 521–534 (2024). https://doi.org/10.1007/s00259-023-06434-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-023-06434-7