Abstract
Objectives
This study investigates the ability of machine learning (ML) models trained on clinical data and 2-deoxy-2-[18F]fluoro-D-glucose(FDG) positron emission tomography/computed tomography (PET/CT) radiomics to predict overall survival (OS), tumor grade (TG), and histologic growth pattern risk (GPR) in lung adenocarcinoma (LUAD) patients.
Methods:
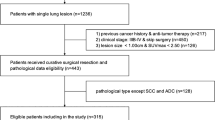
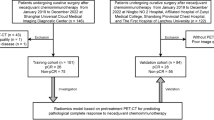
A total of 421 treatment-naive patients with histologically-proven LUAD and available FDG PET/CT imaging were retrospectively included. Four cohorts were assessed for predicting 4-year OS (n = 276), 3-year OS (n = 280), TG (n = 298), and GPR (n = 265). FDG-avid lesions were delineated, and 2082 radiomics features were extracted and combined with endpoint-specific clinical parameters. ML models were built for the prediction of 4-year OS (M4OS), 3-year OS (M3OS), tumor grading (MTG), and histologic growth pattern risk (MGPR). A 100-fold Monte Carlo cross-validation with 80:20 training to validation split was employed as a performance evaluation for all models. The association between the M4OS and M3OS predictions with OS was assessed by the Kaplan-Meier survival analysis.
Results
The area under the receiver operator characteristics curve (AUC) was the highest for M4OS (AUC 0.88, 95% confidence interval (CI) 86.7–88.7), followed by M3OS (AUC 0.84, CI 82.9–84.9), while MTG and MGPR performed equally well (AUC 0.76, CI 74.4–77.9, CI 74.6–78, respectively). Predictions of M4OS (hazard ratio (HR) −2.4, CI −2.47 to −1.64, p < 0.05) and M3OS (HR −2.36, CI −2.79 to −1.93, p < 0.05) were independently associated with OS.
Conclusion
ML models are able to predict long-term survival outcomes in LUAD patients with high accuracy. Furthermore, histologic grade and predominant growth pattern risk can be predicted with satisfactory accuracy.
Key Points
• Machine learning models trained on pre-therapeutic PET/CT radiomics enable highly accurate long-term survival prediction of patients with lung adenocarcinoma.
• Highly accurate survival predictions are achieved in lung adenocarcinoma patients despite heterogenous histologies and treatment regimens.
• Radiomic machine learning models are able to predict lung adenocarcinoma tumor grade and histologic growth pattern risk with satisfactory accuracy.






Similar content being viewed by others
Abbreviations
- CI:
-
95% confidence interval
- FDG:
-
2-deoxy-2-[18F]fluoro-D-glucose
- GLCM:
-
Grey-level co-occurrence matrix
- GLRLM:
-
Grey-level run-length matrix
- GLSZM:
-
Grey-level size zone matrix
- GPR:
-
Histologic growth pattern risk.
- IBSI:
-
Imaging Biomarker Standardization Initiatives
- LUAD:
-
Lung adenocarcinoma
- MC:
-
Monte Carlo
- NGLDM:
-
Neighboring grey-level dependence matrix
- NSCLC:
-
Non-small cell lung cancer
- OS:
-
Overall survival
- TG:
-
Tumor grade
- VOI:
-
Volume of interests
References
Herbst RS, Morgensztern D, Boshoff C (2018) The biology and management of non-small cell lung cancer. Nature 553:446–454
Miller KD, Nogueira L, Mariotto AB et al (2019) Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 69:363–385
Curado MP, Edwards B, Shin HR (2007) Cancer Incidence in Five Continents
Yoshizawa A, Motoi N, Riely GJ et al (2011) Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage i cases. Mod Pathol 24:653–664
Warth A, Muley T, Meister M et al (2012) The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 30:1438–1446
Strand TE, Rostad H, Strøm EH, Hasleton P (2015) The percentage of lepidic growth is an independent prognostic factor in invasive adenocarcinoma of the lung. Diagn Pathol 10:1–7
Li H, Cao W (2020) Pulmonary enteric adenocarcinoma: a literature review. J Thorac Dis 12:3217–3226
Travis WD, Brambilla E, Nicholson AG et al (2015) The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10:1243–1260
Loo PS, Thomas SC, Nicolson MC et al (2010) Subtyping of undifferentiated non-small cell carcinomas in bronchial biopsy specimens. J Thorac Oncol 5:442–447
Soler Cataluña JJ, Perpiñá M, Greses JV et al (1996) Cell type accuracy of bronchial biopsy specimens in primary lung cancer. Chest 109:1199–1203
Huang K-Y, Ko P-Z, Yao C-W et al (2017) Inaccuracy of lung adenocarcinoma subtyping using preoperative biopsy specimens. J Thorac Cardiovasc Surg 154. https://doi.org/10.1016/j.jtcvs.2017.02.059
Ettinger DS, Wood DE, Aggarwal C et al (2019) NCCN Guidelines insights: non-small cell lung cancer, Version 1.2020. J Natl Compr Canc Netw 17:1464–1472
Planchard D, Popat S, Kerr K et al (2018) Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 29:iv192–iv237
Cuaron J, Dunphy M, Rimner A (2013) Role of FDG-PET scans in staging, response assessment, and follow-up care for non-small cell lung cancer. Front Oncol 2:208. https://doi.org/10.3389/fonc.2012.00208
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Papp L, Spielvogel CP, Rausch I et al (2018) Personalizing medicine through hybrid imaging and medical big data analysis. Front Phys 6:1–19
Sha X, Gong G, Qiu Q et al (2019) Identifying pathological subtypes of non-small-cell lung cancer by using the radiomic features of 18F-fluorodeoxyglucose positron emission computed tomography. Transl Cancer Res 8:1741–1749
Shao X, Niu R, Shao X et al (2020) Value of 18F-FDG PET/CT-based radiomics model to distinguish the growth patterns of early invasive lung adenocarcinoma manifesting as ground-glass opacity nodules. EJNMMI Res 10. https://doi.org/10.1186/s13550-020-00668-4
Chang C, Sun X, Wang G et al (2021) A machine learning model based on PET/CT radiomics and clinical characteristics predicts ALK rearrangement status in lung adenocarcinoma. Front Oncol 11:1–12
Kirienko M, Cozzi L, Antunovic L et al (2018) Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging 45:207–217
Valentinuzzi D, Vrankar M, Boc N et al (2020) [18F]FDG PET immunotherapy radiomics signature (iRADIOMICS) predicts response of non-small-cell lung cancer patients treated with pembrolizumab. Radiol Oncol 54:285–294
Kirienko M, Sollini M, Corbetta M, et al (2021) Radiomics and gene expression profile to characterise the disease and predict outcome in patients with lung cancer. Eur J Nucl Med Mol Imaging 48:3643–3655
Edge SB, Compton CC (2010) The american joint committee on cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 17:1471–1474
Travis WD, Brambilla E, Noguchi M et al (2011) International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol 6:244–285
Joo Hyun O, Lodge MA, Wahl RL (2016) Practical percist: a simplified guide to PET response criteria in solid tumors 1.0. Radiology 280:576–584
Stytz MR, Parrott RW (1993) Using kriging for 3d medical imaging. Comput Med Imaging Graph 17:421–442
Zwanenburg A, Leger S, Vallières M, Löck S (2016) Image biomarker standardisation initiative. Radology 295:328–338 https://doi.org/10.1148/radiol.2020191145
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Amin A, Anwar S, Adnan A, et al (2016) Comparing oversampling techniques to handle the class imbalance problem: a customer churn prediction case study. IEEE Access https://doi.org/10.1109/ACCESS.2016.2619719
Huang Y, Liu Z, He L et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non—small cell lung cancer. Radiology 281:947–957
Mu W, Jiang L, Zhang JY et al (2020) Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun 11. https://doi.org/10.1038/s41467-020-19116-x
Yang B, Ji H, Zhong J et al (2020) Value of 18F-FDG PET/CT-based radiomics nomogram to predict survival outcomes and guide personalized targeted therapy in lung adenocarcinoma with EGFR mutations. Front Oncol 10. https://doi.org/10.3389/fonc.2020.567160
Choe J, Lee SM, Do KH et al (2020) Outcome prediction in resectable lung adenocarcinoma patients: value of CT radiomics. Eur Radiol 30:4952–4963
Krajnc D, Papp L, Nakuz TS et al (2021) Breast tumor characterization using [18F]FDG-PET/CT imaging combined with data preprocessing and radiomics. Cancers 13. https://doi.org/10.3390/cancers13061249
Naimi AI, Balzer LB (2018) Stacked generalization: an introduction to super learning. Eur J Epidemiol 33. https://doi.org/10.1007/s10654-018-0390-z
Carvalho S, Leijenaar RTH, Troost EGC et al (2018) 18F-fluorodeoxyglucose positron-emission tomography (FDG-PET)-Radiomics of metastatic lymph nodes and primary tumor in non-small cell lung cancer (NSCLC) – A prospective externally validated study. PLoS One 13:e0192859. https://doi.org/10.1007/s00259-020-04839-2
Bae JM, Jeong JY, Lee HY et al (2017) Pathologic stratification of operable lung adenocarcinoma using radiomics features extracted from dual energy CT images. Oncotarget 8:523–535
Yang SM, Chen LW, Wang HJ et al (2018) Extraction of radiomic values from lung adenocarcinoma with near-pure subtypes in the International Association for the Study of Lung Cancer/the American Thoracic Society/the European Respiratory Society (IASLC/ATS/ERS) classification. Lung Cancer 119:56–63
Funding
This study was funded by the National Major Science and Technology Projects of China (CN) (2016YFC0103705) and the Key Clinical Project of Peking University Third Hospital (BYSYZD2019038).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Weifang Zhang and Xiang Li.
Conflict of interest
L. Papp, M. Hacker, and T. Beyer are co-founders of Dedicaid GmbH, Austria. The remaining authors have no conflict of interest to declare
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
This study was approved by the ethics committee of Peking University Third Hospital (LM2020001).
Methodology
• retrospective
• diagnostic or prognostic study
• observational
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Meixin Zhao and Kilian Kluge shared the first authorship.
Supplementary information
ESM 1
(DOCX 2725 kb)
Rights and permissions
About this article
Cite this article
Zhao, M., Kluge, K., Papp, L. et al. Multi-lesion radiomics of PET/CT for non-invasive survival stratification and histologic tumor risk profiling in patients with lung adenocarcinoma. Eur Radiol 32, 7056–7067 (2022). https://doi.org/10.1007/s00330-022-08999-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08999-7