Abstract
Purpose
To provide a comprehensive assessment of the novel 18 kDa translocator protein (TSPO) radiotracer, [18F]LW223, kinetics in the heart and brain when using a simplified imaging approach.
Methods
Naive adult rats and rats with surgically induced permanent coronary artery ligation received a bolus intravenous injection of [18F]LW223 followed by 120 min PET scanning with arterial blood sampling throughout. Kinetic modelling of PET data was applied to estimated rate constants, total volume of distribution (VT) and binding potential transfer corrected (BPTC) using arterial or image-derived input function (IDIF). Quantitative bias of simplified protocols using IDIF versus arterial input function (AIF) and stability of kinetic parameters for PET imaging data of different length (40–120 min) were estimated.
Results
PET outcome measures estimated using IDIF significantly correlated with those derived with invasive AIF, albeit with an inherent systematic bias. Truncation of the dynamic PET scan duration to less than 100 min reduced the stability of the kinetic modelling outputs. Quantification of [18F]LW223 uptake kinetics in the brain and heart required the use of different outcome measures, with BPTC more stable in the heart and VT more stable in the brain.
Conclusion
Modelling of [18F]LW223 PET showed the use of simplified IDIF is acceptable in the rat and the minimum scan duration for quantification of TSPO expression in rats using kinetic modelling with this radiotracer is 100 min. Carefully assessing kinetic outcome measures when conducting a systems level as oppose to single-organ centric analyses is crucial. This should be taken into account when assessing the emerging role of the TSPO heart-brain axis in the field of PET imaging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Quantification of the 18 kDa translocator protein (TSPO) expression in vivo using positron emission tomography (PET) imaging has proven challenging due to sub-optimal physicochemical properties of previously developed PET radiotracers targeting TSPO (e.g. high non-specific binding and short half-life [1]), complex binding kinetics attributed to the human rs6971 genetic polymorphism [2, 3], and ubiquitous expression of TSPO in mammalian tissues throughout the body [4,5,6,7,8,9]. Consequently, accurate quantification of TSPO PET imaging datasets has predominantly relied on invasive arterial blood collections to generate arterial plasma input function (AIF) data for kinetic modelling.
We have recently developed the TSPO PET radiotracer [18F]LW223, which has binding not susceptible to the human rs6971 genetic polymorphism and is able to detect and quantify macrophage-driven inflammation in a rat myocardial infarction (MI) model and explore the heart-brain axis that exists in this pathology [10]. This radiotracer has a distinct kinetic profile in brain versus heart, and therefore lends itself as an optimal compound for understanding the impact of different binding kinetics on outcome measures.
Although used as the “gold-standard” research method, invasive kinetic analyses are not feasible in routine large-scale clinical nuclear medicine studies. Furthermore, in small animal research, “gold-standard” invasive kinetic modelling is complicated by the limited blood volume of rodents, which requires specific set-ups such as automated blood sampling equipment and surgically induced arterial-venous shunts [11]. This also limits the possibility of performing longitudinal studies which aim to investigate disease progress. To circumvent the issues associated with using AIF for quantification of TSPO PET datasets, simplified methods of quantification have been proposed, including the use of image-derived input functions (IDIF) [12].
We and other colleagues have reported that increased TSPO in the heart due to myocardial infarction begets increased neuronal TSPO expression [5, 13]. The pathological impact of this heart-brain inflammatory axis is yet to be determined but its existence highlights the importance of seeing beyond single-organ centric analysis while pinpointing the need to characterise performance and stability of traditional kinetic models and kinetic constants. Thus far, use of these models and constants is more common in brain PET research, and their application to multiple organs with varying degrees of target expression and radiotracer kinetics has not been as widely explored compared with the use of simplified outcome measures such as SUV.
This paper aims to provide a comprehensive statistical assessment of 18F-LW223 heart and brain kinetic measures when using “gold-standard” AIF versus simplified IDIF analysis protocols of dynamic PET datasets. This is an important component of radiotracer validation, and will also provide a better understanding of some of the complexities associated with analysing the emerging heart-brain axis in the field of TSPO PET imaging.
Materials and methods
Animals and surgical procedures
The data collection and original analysis was described in our previous manuscript [10]. All experiments were authorised by the local University of Edinburgh animal welfare and ethical review committee and in accordance with the Home Office Animals (Scientific Procedures) Act 1986. Fifteen adult male Sprague–Dawley rats (251 ± 4 g) were used. The animals were housed under standard 12 h light:12 h dark conditions with food and water available ad libitum. For MI surgeries, anaesthesia was induced and maintained using isoflurane (0.5–3% in 1.0 l/min oxygen) before buprenorphine (0.05 mg/kg, Alstoe Ltd, York, UK) was administered preoperatively for analgesia. Tracheal intubation was achieved under direct vision, and ventilation was maintained with a rodent ventilator (Harvard Apparatus Model 683, MA, USA, tidal volume 2.5 cm3, respiratory rate 60/min). MI was induced as previously described [14] and MI rats were imaged on 7 ± 1 days post-MI. The naive rat cohort had no surgical procedures and were imaged at a similar timepoint to the MI cohort.
PET/CT imaging
Anaesthesia was induced and maintained with 1.5–2.5% isoflurane (50/50 oxygen/nitrous oxide, 1 l/min). An intravenous (i.v.) line was established in the tail vein for injection of the radiotracer and the femoral artery and vein were cannulated to allow automated blood sample collection, as previously described [11]. The whole blood arterial input function measured by the automatic blood sampler (Swisstrace GmbH, Switzerland) was corrected for the plasma-to-whole blood ratio and for metabolism in vivo (population based), as detailed in our previous manuscript [10]. The average inter-rat variability for the plasma-to-whole blood ratio was 29.3% and for the metabolism ranged between 2.7% (2 min) to 30.9% (60 min). Simulations were carried out on the impact of this level of variability on modelling outputs which demonstrated, unsurprisingly, that a quantitative bias is present (Supplementary Fig. 1). Body temperature was maintained by heated scanner bed or heated mat and monitored by rectal thermometer. Vital signs, including heart rate and respiration rate were monitored continuously during the experiments. PET/CT scans were acquired alongside i.v. bolus injection of [18F]LW223 (23.1 ± 1.7 MBq).
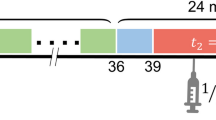
Data were acquired using a PET/CT small animal scanner (nanoPET/CT, Mediso, Hungary). A CT scan (semi-circular full trajectory, maximum field of view, 480 projections, 50 kVp, 300 ms and 1:4 binning) was acquired for attenuation correction. A 120-min emission scan was obtained using 3-dimentional 1:5 mode and re-binned as follows: 18 × 10 s; 2 × 30 s; 1 × 60 s; 2 × 2 min; 10 × 5 min; 6 × 10 min. PET studies were reconstructed using Mediso’s iterative Tera-Tomo 3D reconstruction algorithm, which includes point spread function correction, and the following settings: 4 iterations, 6 subsets, full detector model, low regularisation, spike filter on, voxel size 0.4 mm and 400–600 keV energy window. PET data were corrected for randoms, scatter and attenuation. Ten seconds was the shortest framing possible in this study as the TeraTomo reconstruction algorithm failed to quantitatively reconstruct low count frames less than this.
Image processing and analysis
Reconstructed scans were imported into PMOD version 3.8 (PMOD Technologies, Switzerland). Volumes of interest (VOIs) were manually drawn around the heart and brain using CT images. To sample the infarct area (or equivalent area in naive hearts) averaged PET images (0–120 min) were used to place three spherical VOIs at the centre of the infarct territory within the ventricular wall (1.5 mm3). The IDIF was derived from a VOI placed in the left ventricle of the heart (7 voxel size with each voxel = 0.4 × 0.4 × 0.4 mm) rather than the whole chamber to minimise spill-over influence from the ventricle wall. Time activity curves were generated and standard uptake values (SUVs) calculated as concentration in the VOI divided by injected dose divided by animal weight. Kinetic modelling was performed using the two-tissue compartment model (2TCM) with vB constrained to 0.05 to estimate the kinetic rate constants K1 to k4 and VT in different tissues. 2TCM with time delay corrections of invasive blood data was the most suitable model for the data in these organs based on our previous work, where the full kinetic outcome parameters for the regions used in this study can also be found [13]. The choice of modelling approach and constraints was based on the best compromise between goodness of fitting (AIC) and robustness of fitting (%SE and outcome measure stability). The binding potential relative to non-displaceable volume (BPND) was defined as k3/k4 [15]. The transfer corrected BPND, termed BPTC, was formulated as previously published [13] and as detailed below:
where k3 = radiotracer association rate with specific binding, k4 = dissociation rate constant of target-ligand and K1 = rate constant for transfer from arterial plasma to tissues.
For investigation of the impact of dynamic PET timeframe truncation on modelling outcomes, the 120-min dataset was truncated by 20, 40, 60, 70, and 80 min.
Statistical analysis
GraphPad Prism version 6 (GraphPad Software Inc., USA) was used for statistical analysis and production of graphs. Were appropriate, data are show as the mean ± standard error of the mean (SEM). Pearson correlations were used to determine the strength of associations, with a p < 0.05 considered statistically significant. To assess kinetic outcome measures’ stability, the intraclass correlation coefficient (ICC) was calculated as below:
where \({{\sigma }}_{S}^{2}\) = true variance and \({{\sigma }}_{e}^{2}\) = error variance.
Results
PET outcome measures estimated using AIF and IDIF
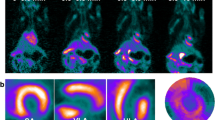
Following MI, there is an upregulation of [18F]LW223 binding within the heart, although in absence of perfusion correction this results in a lower SUV within the infarct (LV anterior wall, Fig. 1a–b). The kinetics of [18F]LW223 in the healthy and infarcted heart display a Morrison’s kinetics binding profile, whereas in the brain the kinetics can be described by the Michaelis–Menten binding principles (Fig. 1b–c) [13].
[18F]LW223 distribution and kinetics in naive rats and rats with myocardial infarction (MI). a Example sagittal SUV images, which are non-perfusion corrected, top row shows the heart and the bottom row the brain. The cardiac areas typically included in the CT drawn global heart VOI are indicated by the broken white line, and the myocardium subsampled within the anterior wall is indicated by a single asterisk (*). A, anterior; P, posterior; H, head and F, foot. b SUV time activity curves for VOIs in the heart and c) brain. Mean ± SEM, n = 6 for naive and n = 9 for MI
The average whole blood time activity curves for AIF and IDIF are shown in Supplementary Fig. 2. Use of IDIF for 2TCM modelling generates outcomes which correlate with those generated using invasive AIF (Fig. 2). Overall, compared to using the invasive AIF, K1 values were 190% higher when using IDIF (based on slope, Fig. 2a–b). The other 2TCM microparameters were higher (k4), lower (k3) and same (k2) when using IDIF (Supplementary Fig. 3). VT and BPTC were 60% (based on slope, Fig. 2c and d) and 90% lower respectively (based on slope, Fig. 2e and f) when using IDIF compared to AIF. This systematic bias when using IDIF is evident within the Bland–Altman plots (Fig. 2b, d and f). When analysing the comparison between AIF and IDIF in naive and MI cohorts on their own, a similar pattern is evident although the fitting is generally poorer within those with MI (Fig. 3).
Comparison of PET outcome measures calculated using the “gold standard” invasive AIF and IDIF in all rats. a Correlation of K1 calculated using AIF vs. IDIF and b Bland–Altman plot for the same comparison. c Correlation of VT calculated using AIF vs. IDIF and d Bland–Altman plot for the same comparison. e Correlation of BPTC calculated using AIF vs. IDIF and f Bland–Altman plot for the same comparison. n = 15 for all graphs (6 naive animals and 9 MI animals) with 3 regions per animal (heart, brain and left ventricular anterior wall)
Comparison of PET outcome measures calculated using invasive AIF and IDIF in separate naive and MI cohorts. a Correlation of K1, b VT and c BPTC calculated using AIF vs. IDIF in naive cohort heart and brain. n = 6 animals with 3 regions (heart, brain and left ventricular anterior wall). d Correlation of K1, e VT and f BPTC calculated using AIF vs. IDIF in the MI cohort heart and brain. n = 9 animals with 3 regions (heart, brain and left ventricular anterior wall)
The impact of truncating PET scan duration on kinetic model outputs
The intraclass correlation coefficients (ICC) of K1, VT and BPTC were calculated for each truncation of the 120-min dynamic PET duration (Fig. 4). K1 ICC values were unaffected by truncations in scan duration (Fig. 4a). The VT ICC demonstrates greater stability in brain outcomes for different truncations compared with heart outcomes (Fig. 4b). The improved brain performance versus the heart maybe due to the decreasing effect of the apparent quasi-irreversible kinetics on 2TCM (Fig. 1). Conversely, for BPTC, a truncation greater than 20 min begins to reduce the number of fittable datasets (defined as the organ data for one animal) and parameter stability in the brain (Fig. 4b and c), but less so in the heart. The kinetic constant values for the hypoperfused left ventricular anterior wall were the most affected by truncation of PET scan duration with the highest number of non-fittable datasets. When assessing the ICC of truncated data within the naive and MI cohorts separately, K1, VT and BPTC ICC results are overall comparable (Supplementary Fig. 4). However, a truncation greater than 20 min still results in loss of fittable datasets.
The ICC of 2TCM parameters for invasive AIF function, IDIF and PET frame truncation in all datasets. a The ICC for K1 calculated using the different conditions in naive and MI rats is shown as dots (left Y axis), with the lines detailing the number of datasets (rats) where calculation of K1 was possible (right Y axis). b The same analysis is shown for VT and c) BPTC. n = 15 (6 naive animals and 9 MI animals)
Furthermore, a truncation of greater than 20 min impacts the quantitative accuracy of VT and BPTC, but not K1, as is evidenced by the deteriorating R2 values and increasing measurement bias (regression line slope, Fig. 5). A similar pattern is seen when assessing the naive and MI cohorts separately (Supplementary Fig. 5, 6 & 7).
The impact of PET scan duration truncation on 2TCM parameter accuracy in all datasets. a Correlation between K1 calculated using a 120-min PET scan duration and 5 other truncated durations. b The same is shown for BPTC and c VT. n = 15 (6 naive animals and 9 MI animals) with 3 regions per animal (heart, brain and left ventricular anterior wall)
Discussion
Here, we investigated bias and stability of [18F]LW223 outcome measures in PET heart and brain imaging from a statistical analysis point of view using “gold-standard” AIF and simplified IDIF analysis protocols. Our results showed that PET outcome measures estimated using IDIF correlate with those derived with invasive AIF, albeit with an inherent bias. This is in agreement with previous observations with other TSPO PET radiotracers [12] as well as other PET studies conducted in rats and mice [16]. This bias is likely due to spill-over issues with IDIF, which deteriorate over time due to radiotracer uptake in surrounding heart tissue. Furthermore, accurate description of a peak image–derived input function (fast perfusion phase) in rodents can be challenging due to the fast heart rate of these animals (c. 300–400 bpm) and limited count rate statistics [17]. VOI placement for IDIF generation is important and the vena cava can be a reliable and reproducible IDIF method for [18F]FDG kinetic modelling in mice, even without partial volume corrections [16, 18]. Albeit, useful in mice PET studies, in rat heart and brain PET experiments, the use of the left ventricle blood pool VOI is more amenable due to limited size of most preclinical PET scanners field of view and larger animal body size. Notwithstanding, because this bias is systematic and strongly correlated with the AIF results, it represents an acceptable trade-off between quantitative accuracy and simplification of protocols, facilitating technology adoption and aiding translational feasibility. Similarly to [18F]FDG metrics that show kinetic modelling provides the most quantitative outcomes [19], large-scale adoption of this radiotracer in the preclinical and clinical settings was pillared on the use of simplified outcome measures like SUV.
The Bland–Altman analysis is a graphical representation, whereby two biomedical techniques can be compared and can be used to assess agreement between two different methods. Baumgartner et al. suggested these plots should be used as a first step in the statistical analysis of radiotracer performance, as they can be useful to understand the dependence of variability on the measured signal and bias of measurements [20]. Our results showed that simplification of protocols (i.e., use of IDIF) resulted in an outcome measure quantitative bias but with a low number of outliers in the Bland–Altman plot; thus, indicating IDIF can produce reliable data.
The variability of PET outcome measures is dependent on organ-specific kinetics, where VT outcomes derived using 120-min data, and AIF were more stable for the brain than the heart tissue. Michaelis–Menten formulations, which have been seamlessly applied to in vivo PET kinetic quantifications, are widely used to model reversible binding kinetic radiotracers [15]. However, in exceptional circumstances, reversibly binding kinetics may also be modelled by the Morrison formulations when there are violations of the free ligand approximation for ligands with high affinity combined with high density targets [21, 22]. TSPO expression in the heart is approximately 5 times higher than in the brain [13, 23, 24], and [18F]LW223 has sub- to nanomolar affinity for TSPO [13]. This results in a considerable fraction of the ligand rapidly binding to the target, thus depleting ligand pool before establishing equilibrium in the heart. In these kinetic circumstances, the macroparameter VT is rate-limited by changes in radiotracer transfer into the tissue poll (K1). Importantly, our data showed that K1 and BPTC had comparable relative variability for all tissues when using 120-min data and AIF, although the preferred outcome measure for the brain was VT (higher ICC values versus BPTC).
According to Baumgartner et al., when describing properties of a given radiotracer, it is important to consider an overall measure across different VOIs and a region-specific measure of reliability in a priori tissue with confirmed biological relevance (e.g. myocardium following infarction). Their work showed that some VOIs had better statistical performance than others, which may be due to different uptake characteristics. Importantly, their study highlighted the utility of comparing statistical performance of the same VOI from different radiotracers as well as different VOIs for the same radiotracer [20]. The work by Baumgartner et al. focused on single-organ (i.e. brain) multi-region analysis and foreseeably these differences of uptake kinetics are likely greater when investigating distinct organs, such as the heart-brain axis. Our results with [18F]LW223 also highlight the importance of validating quantification methods per radiotracer and per organ. For example, the contrasting kinetics in the brain and heart measured with [18F]LW223 in the rat would discourage the use of brain and heart coupling techniques previously proposed to stabilise [18F]GE180 outcome measures in the mouse [25]. It is likely that the differences in organ kinetics with [18F]LW223 PET in rats will also be present in other species, and it should therefore be a factor to consider as research moves into different species towards human translation.
The size of a VOI can also influence noise properties in that region, which in turn can have an impact on variability metrics [20]. This can explain why the left ventricular anterior wall VOI performed worse than the whole heart and whole brain VOI when using 120 min of data and AIF. Although the left ventricular anterior wall VOI placement was conducted in a uniform fashion across studies, the regional territory infarcted following MI surgery may be different depending on affected vessels downstream of the artery ligation. Another limitation of this work is the requirement for a long anaesthesia period (100–120 min), with the need to closely monitor animals to minimise complications such as respiratory depression, particularly across longitudinal studies. Isoflurane may also have a cardioprotective action on the heart, and it has been shown to affect the uptake of other PET radiotracers in the past [26, 27]. In addition, sampling requirements for dynamic IDIF can make accurate description of peak input function challenging, owing to the fast heart rate in small animals and limited count rate statistics.
In conclusion, quantification of [18F]LW223 uptake kinetics in the brain and heart require the use of VT for quantification of TSPO in the brain and BPTC in the hypoperfused heart. This highlights the importance of carefully assessing kinetic outcome measures when conducting different systems level as oppose to single-organ centric analyses. Statistical analysis of [18F]LW223 outcome measures showed the use of simplified IDIF is acceptable and the minimum scan duration for quantification of TSPO expression in rats with this radiotracer and kinetic modelling approach is 100 min. Taken together, these protocol simplifications will enable longitudinal PET imaging studies with this radiotracer in the rat.
Data availability
All data is available on request to the corresponding author. We are currently in the process of adding the data to the digital repository Edinburgh DataShare.
Change history
17 June 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00259-024-06781-z
References
Chauveau F, Boutin H, Van Camp N, Dollé F, Tavitian B. Nuclear imaging of neuroinflammation: a comprehensive review of [11C]PK11195 challengers. Eur J Nucl Med Mol Imaging. 2008;35:2304–19.
Owen DRJ, Gunn RN, Rabiner EA, Bennacef I, Fujita M, Kreisl WC, et al. Mixed-affinity binding in humans with 18-kDa translocator protein ligands. J Nucl Med. 2011;52:24–32.
Owen DR, Yeo AJ, Gunn RN, Song K, Wadsworth G, Lewis A, et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab [Internet]. Nature Publishing Group. 2012;32:1–5. Available from: https://doi.org/10.1038/jcbfm.2011.147
Bird JLE, Izquierdo-Garcia D, Davies JR, Rudd JHF, Probst KC, Figg N, et al. Evaluation of translocator protein quantification as a tool for characterising macrophage burden in human carotid atherosclerosis. Atherosclerosis [Internet]. Elsevier Ireland Ltd. 2010;210:388–91. Available from: https://doi.org/10.1016/j.atherosclerosis.2009.11.047
Thackeray JT, Hupe HC, Wang Y, Bankstahl JP, Berding G, Ross TL, et al. Myocardial Inflammation predicts remodeling and neuroinflammation after myocardial infarction. J Am Coll Cardiol. 2018;71:263–75.
Repalli J. Translocator Protein (TSPO) Role in aging and Alzheimer’s disease. Curr Aging Sci [Internet]. 2015;7:168–75. Available from: http://www.eurekaselect.com/openurl/content.php?genre=article&issn=1874-6098&volume=7&issue=3&spage=168
Abid KA, Sobowale OA, Parkes LM, Naish J, Parker GJM, du Plessis D, et al. Assessing inflammation in acute intracerebral hemorrhage with PK11195 PET and dynamic contrast-enhanced MRI. J Neuroimaging. 2018;28:158–61.
Hatori A, Yui J, Yamasaki T, Xie L, Kumata K, Fujinaga M, et al. PET imaging of lung inflammation with [18F]FEDAC, a radioligand for translocator protein (18 kDa). PLoS One. 2012;7. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0045065
Chelli B, Pini S, Abelli M, Cardini A, Lari L, Muti M, et al. Platelet 18 kDa translocator protein density is reduced in depressed patients with adult separation anxiety. Eur Neuropsychopharmacol. 2008;18:249–54.
MacAskill MG, Stadulyte A, Williams L, Morgan TEF, Sloan NL, Alcaide-Corral CJ, et al. Quantification of macrophage-driven inflammation during myocardial infarction with 18 F-LW223, a novel TSPO Radiotracer with binding independent of the rs6971 human polymorphism. J Nucl Med [Internet]. 2021;62:536–44. Available from: https://doi.org/10.2967/jnumed.120.243600
Warnock G, Bahri MA, Goblet D, Giacomelli F, Lemaire C, Aerts J, et al. Use of a beta microprobe system to measure arterial input function in PET via an arteriovenous shunt in rats. EJNMMI Res [Internet]. Springer Open Ltd; 2011;1:1–11. Available from: http://www.ejnmmires.com/content/1/1/13
MacAskill MG, Walton T, Williams L, Morgan TEF, Alcaide-Corral CJ, Dweck MR, et al. Kinetic modelling and quantification bias in small animal PET studies with [18F]AB5186, a novel 18 kDa translocator protein radiotracer. PLoS One. 2019;14:1–14.
MacAskill MG, Stadulyte A, Williams L, Morgan TEF, Sloan NL, Alcaide-Corral CJ, et al. Quantification of macrophage-driven inflammation during myocardial infarction with 18 F-LW223, a novel TSPO radiotracer with binding independent of the rs6971 human polymorphism. J Nucl Med. 2020;120:243600.
Spath NB, Lilburn DML, Gray GA, Le Page LM, Papanastasiou G, Lennen RJ, et al. Manganese-enhanced T 1 mapping in the myocardium of normal and infarcted hearts. Contrast Media Mol Imaging [Internet]. 2018;2018:1–13. Available from: https://www.hindawi.com/journals/cmmi/2018/9641527/
Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–9.
Lanz B, Poitry-Yamate C, Gruetter R. Image-derived input function from the vena cava for 18F-FDG PET studies in rats and mice. J Nucl Med. 2014;55:1380–8.
Raylman RR, Caraher JM, Hutchins GD. Sampling requirements for dynamic cardiac PET studies using image-derived input functions. J Nucl Med. 1993;34:440–7.
Thackeray JT, Bankstahl JP, Bengel FM. Impact of image-derived input function and fit time intervals on patlak quantification of myocardial glucose uptake in mice. J Nucl Med. 2015;56:1615–21.
Dimitrakopoulou-Strauss A, Pan L, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol Imaging. Eur J Nucl Med Mol Imaging. 2021;48:21–39.
Baumgartner R, Joshi A, Feng D, Zanderigo F, Ogden RT. Statistical evaluation of test-retest studies in PET brain imaging. EJNMMI Res. EJNMMI Research; 2018;8. Available from: https://ejnmmires.springeropen.com/articles/10.1186/s13550-018-0366-8
Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. BBA - Enzymol. 1969;185:269–86.
Williams JW, Morrison JF. [17] The kinetics of reversible tight-binding inhibition. Methods Enzymol [Internet]. 1979;437–67. Available from: https://linkinghub.elsevier.com/retrieve/pii/0076687979630197
Yankam Njiwa J, Costes N, Bouillot C, Bouvard S, Fieux S, Becker G, et al. Quantitative longitudinal imaging of activated microglia as a marker of inflammation in the pilocarpine rat model of epilepsy using [11C]-(R)-PK11195 PET and MRI. J Cereb Blood Flow Metab. 2017;37:1251–63.
Kreisl WC, Fujita M, Fujimura Y, Kimura N, Jenko KJ, Kannan P, et al. Comparison of [11C]-(R)-PK 11195 and [11C]PBR28, two radioligands for translocator protein (18 kDa) in human and monkey: implications for positron emission tomographic imaging of this inflammation biomarker. Neuroimage [Internet]. Elsevier B.V. 2010;49:2924–32. Available from: https://doi.org/10.1016/j.neuroimage.2009.11.056
Deussing M, Blume T, Vomacka L, Mahler C, Focke C, Todica A, et al. Coupling between physiological TSPO expression in brain and myocardium allows stabilization of late-phase cerebral [18F]GE180 PET quantification. Neuroimage [Internet]. Elsevier Ltd. 2018;165:83–91. Available from: https://doi.org/10.1016/j.neuroimage.2017.10.006
Kersten JR, Schmeling TJ, Pagel PS, Gross GJ, Warltier DC. Isoflurane mimics ischemic preconditioning via activation of KATP channels. Anesthesiology [Internet]. 1997;87:361–70. Available from: https://pubs.asahq.org/anesthesiology/article/87/2/361/36186/Isoflurane-Mimics-Ischemic-Preconditioning-via
Hildebrandt IJ, Su H, Weber WA. Anesthesia and Other considerations for in vivo imaging of small animals. ILAR J [Internet]. 2008;49:17–26. Available from: https://doi.org/10.1093/ilar.49.1.17
Funding
This work was funded by the British Heart Foundation (PG/16/12/32022 and PG/17/83/33370) and the Engineering and Physical Sciences Research Council (EPSRC Impact Acceleration Awards, EP/K5039031 and EP/R511705/1). The British Heart Foundation is greatly acknowledged for providing funding towards establishment of the preclinical PET/CT laboratory at the University of Edinburgh (RE/13/3/30183) and radiometabolite laboratories (RG/16/10/32375). AAST, TEFM and MGM are funded by the British Heart Foundation (RG/16/10/32375, FS/19/34/34354). AAST is a recipient of a Wellcome Trust Technology Development Award (221295/Z/20/Z). MGM was also funded by the British Heart Foundation (PG/16/12/32022, PG/17/83/33370). DEN is funded by the British Heart Foundation (CH/09/002, RG/16/10/32375, RE/18/5/34216) and is the recipient of a Wellcome Trust Senior Investigator Award (WT103782AIA). CA-C and CL are supported by the Edinburgh Preclinical Imaging and Edinburgh Imaging core facilities, respectively.
Author information
Authors and Affiliations
Contributions
MGM, DEN, CL, AS, SLP and AAST contributed to conceptualisation of the study. MGM, CW, TEFM, CJA-C and AAST contributed to data generation and analysis. All authors contributed to the preparation of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Conflict of interest
The authors declare the following patent (WO2019243616).
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Cardiology
The original online version of this article was revised: The authors regret that there are some errors in the published original article. All the corrections were listed in the erratum article.
The original article has been corrected.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
MacAskill, M.G., Wimberley, C., Morgan, T.E.F. et al. Modelling [18F]LW223 PET data using simplified imaging protocols for quantification of TSPO expression in the rat heart and brain. Eur J Nucl Med Mol Imaging 49, 137–145 (2021). https://doi.org/10.1007/s00259-021-05482-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-021-05482-1