Abstract
Purpose
[11C]Palmitate PET can be used to study myocardial fatty acid metabolism in vivo. Several models have been applied to describe and quantify its kinetics, but to date no systematic analysis has been performed to define the most suitable model.
Methods
In this study a total of 21 plasma input models comprising one to three compartments and up to six free rate constants were compared using statistical analysis of clinical data and simulations. To this end, 14 healthy volunteers were scanned using [11C]palmitate, whilst myocardial blood flow was measured using H2 15O.
Results
Models including an oxidative pathway, representing production of 11CO2, provided significantly better fits to the data than other models. Model robustness was increased by fixing efflux of 11CO2 to the oxidation rate. Simulations showed that a three-tissue compartment model describing oxidation and esterification was feasible when no more than three free rate constants were included.
Conclusion
Although further studies in patients are required to substantiate this choice, based on the accuracy of data description, the number of free parameters and generality, the three-tissue model with three free rate constants was the model of choice for describing [11C]palmitate kinetics in terms of oxidation and fatty acid accumulation in the cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The myocardium relies on several sources of energy, including polysaccharides (sugar) and short and long fatty acids. Measuring kinetics of these compounds can give valuable information about myocardial function under various conditions [1, 2]. Uptake and utilization of polysaccharides can be measured using FDG. The kinetic behaviour of FDG is well understood and can be described accurately by an irreversible two-tissue compartmental model [2, 3]. For studying oxidation of other energy sources (MVO2) of the heart, the short-chain fatty acid [11C]acetate is often used [4, 6, 7]. Although kinetics of this tracer were originally studied using a six-compartment model, the limited time resolution of a PET study forced a reduction to a simplified two-tissue compartment model describing uptake (and back diffusion) and (irreversible) oxidation of the tracer [6, 7].
Finally, [11C]palmitate can be used to study uptake and utilization of long-chain fatty acids, which play a dominant role in delivering energy to the heart. Since fatty acid metabolism [8] is affected by a number of pathophysiological conditions, [11C]palmitate has been used to study myocardial metabolism in various diseases such as viral infections and diabetes [5, 10]. Most studies involving [11C]palmitate used semiquantitative analyses to describe its kinetics. Tracer washout is commonly described using a biexponential function with a rapid phase reflecting fast oxidation of fatty acid in the mitochondria and a slower phase, appearing later, representing incorporation of extracted fatty acids into neutral lipid pools [10]. However, a simpler single-exponential model, only describing oxidation of the tracer has also been used [11].
A possibly more accurate three-tissue compartmental model (hence described by three exponentials), in which uptake of palmitate in cytosol, beta-oxidation in mitochondria and incorporation of fatty acid into neutral lipids is described, has been proposed by Bergmann et al. [9]. The model was validated using dogs under several conditions and subsequently used to assess altered myocardial fatty acid metabolism associated with cardiomyopathy in patients [12]. Although the proposed model corresponds with known pathways of fatty acids and therefore is a valid model, to date no details about accuracy, reproducibility or identifiability of individual pathways have been reported.
In the present study, simulations and actual PET data from healthy controls were used to evaluate the above-mentioned three-tissue compartment model, together with other, simplified, models describing [11C]palmitate kinetics. Although final model choice will also depend on physiological relevance, parameter identifiability of the model is a first and mandatory criterion in model choice and was the overall aim of this study.
Materials and methods
Patients
A group of 14 healthy men (Table 1), aged 53.7±6.0 years, were recruited by advertisements in local newspapers. Healthy men without complaints or any history of cardiovascular disease were eligible. Inclusion criteria were blood pressure not exceeding 140/90 mmHg, body mass index <32 kg/m2, and normal glucose tolerance. Exclusion criteria were any clinically significant disorder, dyslipidaemia, requiring pharmacological treatment, or the use of any prescribed drug. Subjects were screened, including history, physical examination, echocardiogram, and fasting blood and urine analyses. Normoglycaemia was determined using a 75-g oral glucose tolerance test. Written informed consent was obtained from all participants. The protocol was approved by the local Medical Ethics Committee, and performed in full compliance with the Declaration of Helsinki.
Production of PET tracers
[11C]Palmitate was synthesized according to a modified method previously described by Welch et al. [13] and using nuclear interface equipment. Radiochemical purity was >97%.
Study procedures
Subjects arrived at the PET facility at 08.00 a.m. after an overnight fast. They had been instructed to abstain from alcohol, caffeine-containing beverages and physical exercise for 48 h prior to PET scanning. For six subjects arterial and venous samples taken during the [11C]palmitate scan were compared with regard to specific activity concentrations including metabolites. Since no significant difference was found in these blood values all other subjects received only an indwelling venous catheter in an antecubital vein. During all procedures patients were monitored by telemetry and blood pressure was measured at 5-min intervals. Blood samples were collected during both scans at predefined intervals to measure plasma substrate (glucose, nonesterified fatty acids, lactate, triglycerides) and insulin levels. In addition samples for plasma/whole-blood ratios and 11CO2 concentrations were drawn and analysed immediately.
PET scanning
All scans were performed using an ECAT EXACT HR+ scanner capable of 2-D and 3-D acquisition (Siemens/CTI, Knoxville, TN) [14, 15]. A 10-min transmission scan, using three rotating 68Ge line sources, together with sinogram windowing, was performed after subject positioning at the cardiac level. Next, to measure myocardial blood flow (MBF), 1,100 MBq H2 15O dissolved in 5-ml saline was injected intravenously as a bolus, followed by a 40-ml saline flush at a rate of 4 ml/s. A dynamic emission scan was acquired consisting of 40 frames with variable frame length for a total duration of 10 min (12×5, 12×10, 6×20, and 10×30 s). This scan was performed in 2-D mode to prevent large dead time effects at the beginning of the scan [16]. After 10 min to allow for decay of residual 15O, fatty acid imaging was performed using [11C]palmitate. An intravenous bolus of 180 MBq was followed immediately by a 30-min dynamic emission scan in 3-D mode for increased sensitivity. This scan was divided into 33 frames (6×5, 6×10, 3×20, 5×30, 5×60, and 8×150 s). All emission data were corrected for physical decay, dead time, scatter, randoms, and (measured) photon attenuation. Image reconstruction was performed using filtered back projection (FBP) with a Hanning filter at 0.5 of the Nyquist frequency, resulting in a spatial resolution of about 7 mm full-width at half-maximum. In addition a 10 to 30-min sum image was reconstructed with OSEM.
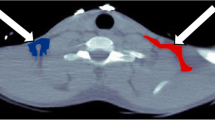
In order to generate myocardial time–activity curves (TACs), regions of interest (ROIs) were defined on resliced short-axis [11C]palmitate images using the OSEM reconstructed summed image from 10 to 30 min. Six basal ROIs, six distal ROIs and one apical ROI were defined as described previously [17]. Additional ROIs were defined in left ventricular chambers for image-derived input functions (IDIF). An example of ROI placement is shown in Fig. 1. Myocardial segments exposed to liver spill-in were omitted from the analysis of [11C]palmitate scans, whilst all other ROIs were grouped for further analysis. All ROIs were projected onto the dynamic H2 15O image data to generate water TACs.
Plasma input
An [11C]palmitate plasma input function was generated by multiplying the image-derived left ventricle TAC by the plasma to whole blood ratio and by correcting for the fraction of 11CO2 as obtained from the venous samples. As these corrections were based on a limited number of manually drawn blood samples, spline interpolation was used to correct values at every time-frame of the PET scan.
Kinetic models
Myocardial blood flow
H2 15O data were fitted to a standard four-parameter, single-tissue compartment model including MBF, perfusable tissue fraction and spill-over fractions from both ventricles [18]. The left ventricular cavity TAC was used as input function to the model.
[11C]Palmitate fatty acid metabolism
A total of 21 different models with different combinations of compartments, rate constants and constraints were investigated. The models were divided into four families depending on the number and type of compartments that were included. Within a family of models, the number of rate constant and constraints were varied (Table 2).
For family 1, one compartment was used to describe uptake of [11C]palmitate from plasma into tissue (kp1), and washout from tissue back to plasma (k1p). This model does not distinguish between back diffusion of [11C]palmitate or efflux of 11CO2. Family 2 included both a free or interstitial compartment and a compartment describing turn-over of free fatty acids into neutral lipids and amino acids, with rate constants kp1, k1p, k12, k21 describing transfer rates between compartments. In one implementation (model 2B), the back-diffusion process (k21) was omitted, limiting the total number of free parameters to three.
Although also constructed from two compartments, models from family 3 differed from family 2 models in terms of metabolic pathways. Instead of (slow) turn over of free fatty acids, it included a mitochondrial compartment with k13 describing transport of [11C]palmitate into the mitochondria and k3p efflux of 11CO2 into the blood. Different implementations were obtained by omitting k1p (model 3B), whilst further reduction of the number of free rate constants was obtained by fixing k3p to a value proportional to flow (MBF/Vb, with Vb=0.1, being the blood volume of the myocardium; models 3A(1) and 3B(1)) or by imposing k3p=k13 (models 3A(2) and 3B(2)).
The largest family of models (family 4) included all three compartments introduced by Bergmann et al. [9]. Reduction of the number of free parameters was accomplished by omitting either or both k1p and k21 (models 4C, 4B and 4D, respectively). In addition, further modifications (reductions) were obtained by fixing k3p in a similar manner as for family 3. Using family 4 models, myocardial fatty acid uptake (MFAU), myocardial fatty acid oxidation (MFAO) and myocardial fatty acid esterification (MFAE) can be estimated separately (see below).
Implementation
For every model a set of differential equations can be defined. Its analytical solution can then be fitted to myocardial PET data, yielding estimates of the various parameters. As all models can be deduced from the description of model 4A by setting specific parameters to zero (or to fixed values), only the solution of model 4A is given. Similar to the suggestion of Degrado et al. [19], this model can be solved and described by three exponentials [19]:
C PET (t) is the measured [11C]palmitate TAC, C p (t) the plasma input, C lv (t) the blood TAC taken from the left ventricle, S lv the unknown spill-over of arterial signal into the myocardium, and k 1p , k p1 , k 12 , k 21 , k 13 and k 3p are the rate constants as described above. Since spill-over from the right ventricle was found to be negligible, it is not included in the model. The fraction of tissue TAC representing real myocardial tissue activity (perfusable tissue fraction, PTF) cannot be estimated independently from k p1 and was therefore estimated as (1−S lv ). This was based on the finding that this specific relationship between the spill-over factor S lv and PTF was present in the H2 15O studies and the assumption that the underlying physics determining these factors (myocardial wall movement, vascularity, limited resolution) were comparable in both studies. The macroparameters MFAO and MFAE were defined according to Bergmann et al. [9]:
and MFAU equals the sum of MFAO and MFAE. C NEFA is the concentration of nonesterified free acids in blood (in millimoles per gram). In brief, fitting Eq. 1 to the [11C]palmitate data directly yielded the parameters k p1 … k 3p , from which the macroparameters could be calculated. When applying models that did not include backflow from compartment 1 to plasma, k 1p was set to zero in Eqs. 2 and 3.
Model analysis
All models were fitted to C PET (t) using a nonlinear least-squares optimization process based on the Levenberg-Marquart algorithm [20]. Results were analysed using the following criteria:
Residual errors
The residual or sum of squared errors was used to describe how well the estimated curve fitted to the data. Values averaged over all subjects are reported.
Akaike information criterion (AIC)
Similar to the residual errors, except that penalties are given for the number of parameters used. AIC includes the effect that models with a larger number of free parameters are more likely to find solutions that are closer to the data, but yield parameter estimates with lower confidence.
Average parameter values
For each model, average values and standard deviations of each parameter were determined over the entire group of normal subjects. As it concerns a relatively homogeneous group of volunteers, the standard deviation, SD, of the distribution of values within the group is some indication of the precision of the estimate. Furthermore, the standard error, SE, in parameter estimation was calculated based on the Jacobian that results from the fitting procedure for each subject and parameter. SE is a measure of robustness of the parameter estimation. SE values averaged over all subjects are reported.
Sensitivity analysis
The relative change in model outcome or TAC resulting from a change in an individual parameter is indicative of how accurately the parameter can be estimated. Sensitivity functions for each parameter were generated by determining the change in model outcome resulting from a 1% change in the parameter [21, 22]. First, typical arterial TACs (C p (t) and C lv (t)) were generated by averaging the measured curves over all studies. These were then used to generate tissue TACs. If each of the sensitivity functions for the parameters are sufficiently different over time, then the parameters are identifiable. To test the ability of the model to estimate parameters independently, we used identifiability analysis of the sensitivity curves to generate a sensitivity matrix as previously described [21, 22]. Each element in the sensitivity matrix represents the correlation between any two parameters to produce a change in the output of the model. Pairs of parameters within the correlation matrix elements close to ±1 will have estimates that covary considerably and are therefore difficult to estimate independently. Furthermore, the required length of the experimental data collection and analysis may be indicated from the sensitivity functions. To evaluate the impact of lengthening the scan duration, sensitivity curves and correlation matrices were also generated for a 45-min scan.
Monte Carlo simulations
These were performed to assess accuracy of estimated MFAU, MFAE and MFAO over a wide range of parameter values. For every model a set of 100 tissue TACs was generated using randomly selected parameters. Simulations were performed with and without addition of Gaussian noise. In reality, the noise level in the ROI depends on many factors including ROI, acquisition and reconstruction parameters and cannot be measured directly due to unknown real activity distribution and local heterogeneity. Therefore, noise level was based on total (non-decay corrected) trues T per frame, with a coefficient of variation equalling α/√T. For all simulations α was chosen such that the residuals of the model fits to the simulated data were similar to those found in the patient data using the most precise model (4A), which is the best estimate of the true TAC. Subsequently the simulated tissue TACs were fitted to the models to yield estimated parameters. Since this was done over the full range of parameter values, scatter plots of true vs. estimated parameter values could be generated, illustrating the confidence of the estimated parameter. For every simulation the model used for parameter estimation was the same model as that used for generating the TACs. Ideally, for a noiseless situation and a suitable model, a perfect correlation between simulated and estimated parameters is to be expected.
Results
Myocardial blood flow
MBF values averaged 0.81±0.21 ml/ml per minute. In relevant models (Table 2), these MBF values were used to fix the outflow of 11CO2 from mitochondria to the vascular space described by parameter k 3p .
[11C]Palmitate
In Fig. 2 (top) a typical example is shown of a correction curve (based on manual sample data) used to translate the image-derived whole-blood curve into a plasma input curve that is corrected for differences in plasma and whole-blood concentrations and for the presence of 11CO2. A myocardial TAC with associated fits from a variety of models is also depicted in Fig. 2. For all models, the average left ventricular spill-over fraction S lv was 0.18±0.05. A high correlation (R 2=0.998, data not shown) was found between spill-over factors estimated with different models.
Top Example of a typical correction curve (together with measured manual sample data) for transferring an image-derived whole-blood curve into a plasma input curve corrected for the presence 11CO2. Bottom Example of myocardial [11C]palmitate data (dots) together with best fits (lines) using various models
Residual errors
Table 3 shows average residual errors of all models included in the study. In general, the error decreased with increasing number of free parameters. As compared to the single-compartment model (family 1), inclusion of the mitochondrial compartment (compartment 3) improved the quality of the fits dramatically (families 3 and 4), except when k 3p was fixed to MBF/V b . This improvement was not seen when only the compartment representing the slow turnover pool was included (family 2). Lowest residual errors were obtained when all three compartments were included (family 4), although differences between families 3 and 4 were very small. Fixing k 3p to a value proportional to MBF (i.e. to MBF/V b ) resulted in much larger errors than fixing it to k 13 .
Akaike Information Criterion
In Table 3 the preference for each model based on AIC values is also given. In addition to the residual error, AIC also takes into account the number of free parameters in the model and therefore it can be regarded as a more sensible measure of goodness of fit. In 50% of the subjects, model 3B(2) was the preferred model.
Average parameter values
Table 4 shows average values and standard deviations (SD) of the various parameters across the subjects for most models. In addition, mean standard errors (SE) of the fitted parameters are provided. As residual error and AIC analyses had already indicated preference for models in which k 3p was fixed to k 13 , models in which k 3p was fixed to MBF/V b were excluded from further analysis. From Table 4 it can be seen that k p1 can be estimated with high precision, nearly independent of model choice. For all other parameters, actual parameter values (and spread, i.e. both SD and SE) heavily depended on model choice. This is especially the case for k 21 and k 12 , and to a lesser extent, for k 13 . In concordance with this, all models that consisted of more than three free rate constants yielded at least one parameter with an SE that was an order of magnitude larger than the average parameter value.
Sensitivity analysis
As input for the sensitivity analysis, average clinical parameters were used together with average curves for plasma input, and left and right ventricles. Figure 3 shows sensitivity curves for four models (3B(2), 4D(2), 4C(2) and 4A(2)). Since the curves of the various parameters of models 3B(2) and 4D(2) differed significantly, it was expected that these parameters could reliably be estimated independently. For models 4A(2) and 4C(2), however, the shape of the curves for parameters k 1p , k 12 , k 21 and k 13 were very similar. Correlation values between parameters are given in Table 5. For model 4A(2), there was a high level of correlation between all parameters except k p1 . This suggests that it would be difficult to obtain independent estimates of these parameters using model 4A(2). Similarly, k 21 and k 13 were highly correlated with k 12 for model 4C(2). In contrast, for model 4D(2) all correlations were smaller than 0.9, yielding better conditions to independently estimate its parameters, although the independence of k 13 remains somewhat dubious in this analysis. Also shown in Table 5 are the correlation values for a 45-min scan. Increasing the scan length by 50% had a positive effect on the estimation of k 13 using the 4D(2) model only.
Monte Carlo simulations
As input, average blood curves as obtained from the clinical study were used. Scatter plots are given in Fig. 4 for noiseless curves. From the scatter plots, Pearson correlation coefficients were calculated for simulations both without and with gaussian noise (Table 6). For all models, parameter k p1 and changes in MFAU could be detected with high accuracy. When using model 4C(2) and especially 4A(2), significant uncertainties in MFAO estimation were found even in the noise-free situation. MFAE could only be estimated reliably using model 4D(2), while 4C(2) again yielded large uncertainties. In addition, using model 4A(2) MFAE contained large biases. Interestingly, the effects of noise on the confidence of parameter estimate were small, probably due to the use of large ROIs (Table 6).
Direct comparison of models 3B(2) and 4D(2)
In order to investigate to what extent a three-tissue compartment model could be used for analysing data, a direct comparison was made between two-tissue model 3B(2) and three-tissue model 4D(2). The first model was the most preferred amongst all models in the AIC analysis, whilst the latter was preferred amongst models of family 4. In addition, model 4D(2) was the only model that showed promising results in the sensitivity and Monte Carlo analyses. Again, using AIC, a direct comparison was made between models 3B(2) and 4D(2) across all 14 subjects. According to the AIC, model 3B(2) was preferred in eight subjects and model 4D(2) in six (Fig. 5). Hence, the total population was split into a ‘3B(2) group’ with eight subjects and a ‘4D(2) group’ with six subjects. As well as this, all 14 subjects were analysed with model 4D(2), so that for every subject in both groups a k 12 value was found. For the ‘3B(2) group’ the average value of k 12 was 0.005±0.005 min−1. In contrast, for the ‘4D(2) group’ it was 0.024±0.013 min−1 (Fig. 5). Hence, as could have been expected, model 4D(2) appeared to be the model of choice when k 12 is relatively high. Importantly, using model 4D(2) for all subjects had negligible effects on the estimation of k p1 (R 2=0.999) and k 13 (R 2=0.989), as shown in Fig. 5.
Top left Direct comparison between models 3B(2) and 4D(2) in terms of preferred model according to the AIC. Top right Average k 13 as determined using model 4D(2) for the groups of volunteers in which models 3B(2) and 4D(2) were preferred. Bottom Correlation between k p1 values and between k 13 values, both as obtained with models 3B(2) and 4D(2)
Discussion
This study addressed possible kinetic models and their potential to describe uptake of [11C]palmitate in the myocardium. The suitability of a kinetic model depends on several factors. Based on biochemical pathways of the tracer, as known from in vitro or animal studies, kinetics can be expressed adequately by more comprehensive tracer compartment models. For [11C]palmitate, such a model typically comprises ten compartments with associated rate constants, which clearly would provide an adequate description of tracer kinetics. In order to use a model in PET studies, however, it is usually necessary to simplify the compartmental structure to meet the limited information contained within dynamic PET data. Ideally, but not possible in human studies, the outcome of a model should then be compared with direct measurement of the kinetics at the organ or cell level. As both physiological description and clinical consequences of [11C]palmitate model choice have previously been discussed in detail [9, 11, 23, 24], the present study focused on the applicability of these models within spatial and temporal limitations of the PET data. To assess how many parameters could be determined from human [11C]palmitate studies, models of different levels of complexity were compared systematically, in each case relating the model to the underlying physiology [25–27].
Previously, a three-tissue compartment model has been proposed, describing uptake, oxidation and esterification of [11C]palmitate in the myocardium, which was evaluated in dogs [9]. Although it is well known that actual metabolic pathways are much more complex, it would in theory be reasonable to describe the various pathways using this simplified model, given the limited kinetic information of the data. However, even a three-tissue compartment model with a relatively large number of fit parameters (up to seven) can be challenging. As a matter of fact, to our knowledge, no other PET tracer kinetic model with so many parameters has successfully been used before. Therefore, in the present study, both PET data from volunteers and simulated data were used to investigate whether a three-tissue compartment model could be used in practice. As potential alternatives to the three-tissue compartment model, both single and two-tissue (i.e. further simplified) compartment models were included in order to perform a formal assessment of the number of identifiable compartments.
Since there is no gold standard for in vivo determination of fatty acid metabolism in the heart, no direct accuracy of model outcome could be determined. Some data on fatty acid metabolism in the heart has been published however, using a more simplified biexponential model [10]. For comparison, the patient data in this study were also analysed with the biexponential model: average uptake indices (±SD) were 0.08±0.02 vs. 0.11±0.01, and clearance half-times were 19.1±3.1 min vs. 17.6±1.6 min for this study and the study by Knuuti et al. [10], respectively.
Preferred model
Although a number of mathematical methods were applied to investigate applicability of models, their conclusions were concordant. AIC and standard error analysis both showed that models including more than three free rate constants yielded parameters with very low confidence, which was due to strong interparameter correlation, as indicated by the sensitivity analysis. Monte Carlo analysis finally confirmed this finding by showing that noise had little effect on parameter identifiability.
Sources of error
Simplified models with a limited number of free rate constants are likely to be more robust for parameter estimation, but they also yield less information about underlying physiology. For example, the outcome of a single-tissue compartment model is restricted to influx and egress of [11C]palmitate and does not provide a separate assessment of the oxidative pathway, possibly obscuring sources of altered fatty acid metabolism. Although unlikely, 11CO2 itself could hypothetically be taken up from blood into tissue. This could be investigated using 11CO2 as a primary tracer [28]. Furthermore, additional studies are needed to assess whether other metabolites are produced and, if so, how sensitive the model is to the presence of these metabolites.
For the clinical data possible sources of error were similar to those in PET studies in general. Although quantification of PET data is never a matter of course, scanner and reconstruction methods used in this study were well validated and can be regarded as proven technology. To counteract the effect of limited resolution and cardiac wall movement, spill-over and perfusable tissue factors were included in the model. Furthermore, all studies were manually checked by comparing the position of the myocardium through the dynamic which revealed no patient movement. The impact of noise on the analysis was minimized by using summed ROIs. The Monte Carlo simulations included a similar noise level to that in the clinical data, and showed no large detrimental influence of noise.
For the simulations a possible source of error could have been a physiologically incorrect model or wrong choice of parameters. In order not to give undue preference to a model, for the Monte Carlo simulated data the same model was used as for the parameter estimation of these data (matched models), making the clinical validity of the model less relevant. Hence, errors in simulated parameter estimation would have come from either simulated noise or interdependency of variables. The average parameter values used for simulation were based on the clinical results. Since these were unreliable for some models and could have varied significantly, a range of values was used in the scatter analysis to try to include all possible situations.
Oxidation and esterification
The major finding from the present systematic model analysis is that an oxidative pathway should be incorporated when describing [11C]palmitate metabolism. This is underscored by the fact that model 3B(2), a two-tissue compartment model with free parameters k p1 and k 13 , was preferred over the single-tissue compartment model 1 with free parameters k p1 and k 1p , although both models had an equal number of free parameters. In order to reduce the number of free parameters, Bergmann et al. proposed fixing k 3p to MBF/V b that could be derived from an independent MBF measurement. Although the found MBF values are in concordance with the generally found average and range of MBF values [29], in the present study such an approach did not yield optimal fits. This could have been due to incorrect assumptions about the value of V b . However, no correlation between MBF as determined with H2 15O and k 3p as determined with models 4A, 4B, 4C or 4D (best correlation R 2=0.28) could be found, suggesting that if k 3p is equal to MBF/V b , V b itself is not constant. Therefore physiological variation in V b , or even a coupling between MBF and V b , could be expected. Hence, although in principle, incorporating additional information to the model could increase reliability, the present results raise doubts as to whether MBF/V b with a constant V b could be used to independently determine k 3p , at least in healthy human subjects.
Monte Carlo simulations suggested that the metaparameters describing uptake, oxidation and esterification could be estimated accurately using model 4D(2), the simplest three-tissue compartment model. Analyses using model 4C(2) indicated that adding just one additional free parameter (compared with model 4D(2)) significantly decreased the confidence of metaparameter estimation. This is illustrated by the spread seen in Fig. 4 for model 4C(2), where ideal conditions were used (i.e. no noise in the simulated data). The limited value of all models of family 4 other than model 4D(2) is underlined by the sensitivity analyses that showed large correlations between different parameters even when the scan length was increased by 50% (Table 5).
Although model 4D(2) could be used to describe [11C]palmitate kinetics, in a small majority of all subjects (8 out of 14, 57%) model 3B(2) was preferred, probably because parameter k 12 was negligible in this group, and hence, there was no added value in applying model 4D(2). This is in agreement with the general situation that free fatty acid in healthy subjects is primarily oxidized rather than accumulating in the myocardial cell. Although there is no direct argument for using the three-tissue compartment model for this specific group, the comparison between models 3B(2) and 4D(2) showed that estimated values of the other parameter were hardly affected, and therefore it could still be used without loss of precision. On the other hand, if model 3B(2) were to be used for the group in which model 4D(2) was preferred, esterification wrongly could be ignored according to the AIC.
An important application of [11C]palmitate will be in subjects with heart disease with possibly altered metabolism as compared to the subjects with healthy aerobic hearts included in this study. It is likely that the esterification pathway is even more pronounced in some patient groups, such as patients with type 2 diabetes mellitus. In such patients, the use of model 4D(2) is recommended, as it allows for esterification without compromising the results when this pathway is negligible.
Conclusion
The optimal model for analysing [11C]palmitate data is a three-tissue compartment model in which three rate constants need to be fitted. Inclusion of an oxidative pathway in the model is essential and best fits were obtained by fixing the efflux rate of 11CO2 to the oxidation rate. Inclusion of a (slow) esterification compartment had less impact, improving fits in about half of the normal subjects. Nevertheless, inclusion of this compartment is recommended as it had only a minor impact on the reliability of the other parameters, whilst allowing for more pronounced esterification, which is likely to be present in some patient groups.
References
Miyabe H, Ohte N, Iida A, Narita H, Yoshida T, Kimura G. Evaluation of fatty acid beta-oxidation in patients with prior myocardial infarction in relation to myocardial blood flow, total oxidative metabolism, and left ventricular wall motion. Circ J. 2005;69:1459–65.
Phelps ME, Mazziotta JC, Schelbert HR. Positron emission tomography and autoradiography (Principles and applications for the brain and heart). New York: Raven; 1986.
Sokoloff L, Reivich M, Kennedy C, Des Rosiers MH, Patlak CS, Pettigrew KD, et al. The [14C]deoxyglucose method for the measurement of local cerebral glucose utilization: theory, procedure, and normal values in the conscious and anesthetized albino rat. J Neurochem. 1977;28:897–916.
Klein LJ, Visser FC, Knaapen P, Peters JH, Teule GJJ, Visser CA, et al. Carbon-11 acetate as a tracer of myocardial oxygen consumption. Eur J Nucl Med. 2001;28:651–68.
Schelbert HR, Henze E, Sochor H, Grossman RG, Huang SC, Barrio JR, et al. Effects of substrate availability on myocardial C-11 palmitate kinetics by positron emission tomography in normal subjects and patients with ventricular dysfunction. Am Heart J. 1986;111:1055–64.
Sciacca RR, Akinboboye O, Ling Chou R, Epstein S, Bergmann SR. Measurement of myocardial blood flow with PET using 1-11C-acetate. J Nucl Med. 2001;42:63–70.
Sun KT, Yeatman LA, Buxton DB, Chen K, Johnson JA, Huang SC, et al. Simultaneous measurement of myocardial oxygen consumption and blood flow using [1-carbon-11]acetate. J Nucl Med. 1998;39:272–80.
van der Vusse GJ, Glatz JF, Stam HC, Reneman RS. Fatty acid homeostasis in the normoxic and ischemic heart. Physiol Rev. 1992;72:881–940.
Bergmann SR, Weinheimer CJ, Markham J, Herrero P. Quantitation of myocardial fatty acid metabolism using PET. J Nucl Med. 1996;37:1723–30.
Knuuti J, Takala TO, Nagren K, Sipila H, Turpeinen AK, Uusitupa MI, et al. Myocardial fatty acid oxidation in patients with impaired glucose tolerance. Diabetologia 2001;44:184–7.
Goldstein RA, Klein MS, Welch MJ, Sobel BE. External assessment of myocardial metabolism with C-11 palmitate in vivo. J Nucl Med. 1980;21:342–8.
Bergmann SR, Herrero P, Sciacca R, Hartman JJ, Rubin PJ, Hickey KT, et al. Characterization of altered myocardial fatty acid metabolism in patients with inherited cardiomyopathy. J Inherit Metab Dis. 2001;24:657–74.
Welch MJ, Dence CS, Marshall DR, Killbourn MR. Remote system for production of carbon-11 labelled palmitic acid. J Lab Comp Radiopharm. 1983;20:1087–95.
Brix G, Zaers J, Adam LE, Bellemann ME, Ostertag H, Trojan H, et al. Performance evaluation of a whole-body PET scanner using the NEMA protocol. J Nucl Med. 1997;38:1614–23.
Adam LE, Zaers J, Ostertag H, Trojan H, Bellemann ME, Brix G. Performance evaluation of the whole-body PET scanner ECAT exact HR+ following the IEC standard. IEEE Trans Nucl Sci. 1997;44:1172–9.
De Jong HWAM, Lubberink M. Issues in quantification of cardiac PET studies. Eur J Nucl Med Mol Imaging. 2007;34:316–9.
Knaapen P, Boellaard R, Gotte MJW, Dijkmans PA, van Campen LMC, de Cock CC, et al. Perfusable tissue index as a potential marker of fibrosis in patients with idiopathic dilated cardiomyopathy. J Nucl Med. 2004;45:1299–304.
Hermansen F, Rosen SD, Fath-Ordoubadi F, Kooner JS, Clark JC, Camici PG, et al. Measurement of myocardial blood flow with oxygen-15 labelled water: comparison of different administration protocols. Eur J Nucl Med. 1998;25:751–9.
Degrado TR, Holden JE, Ng CK, Raffel DM, Gatley SJ. Quantitative-analysis of myocardial kinetics of 15-P-[Iodine-125] iodophenylpentadecanoic acid. J Nucl Med. 1989;30:1211–8.
More JJ. The Levenberg-Marquardt algorithm: implementation and theory, numerical analysis. In: Watson GA, editor. Lecture notes in mathematics 630. Springer Verlag; 1977. pp. 105–116.
Muzi M, Mankoff DA, Grierson JR, Wells JM, Vesselle H, Krohn KA. Kinetic modeling of 3′-deoxy-3′-fluorothymidine in somatic tumors mathematical studies. J Nucl Med. 2005;46:371–80.
Mankoff DA, Shields AF, Graham MM, Link JM, Eary JF, Krohn KA. Kinetic analysis of 2-[carbon-11]thymidine PET imaging studies: compartmental model and mathematical analysis. J Nucl Med. 1998;39:1043–55.
Arai T, Wakabayashi SI, Channing MA, Dunn BB, Der MG, Bell JM, et al. Incorporation of [1-carbon-11]palmitate in monkey brain using PET. J Nucl Med. 1995;36:2261–7.
Geltman EM. Assessment of myocardial fatty acid metabolism with 1-11C-palmitate. J Nucl Cardiol. 1994;1:S15–22.
Gunn RN, Sargent PA, Bench CJ, Rabiner EA, Osman S, Pike VW, et al. Tracer kinetic modeling of the 5-HT1A receptor ligand [carbonyl-11C]WAY-100635 for PET. Neuroimage 1998;8:426–40.
Lammertsma AA. Radioligand studies: imaging and quantitative analysis. Eur Neuropsychopharmacol 2002;12:513–6.
Kropholler MA, Boellaard R, Schuitemaker A, Folkersma H, van Berckel BNM, Lammertsma AA. Evaluation of reference tissue models for the analysis of [11C](R)-PK11195 studies. J Cereb Blood Flow Metab. 2006;26:1431–41.
Brooks DJ, Lammertsma AA, Beaney RP, Leenders KL, Buckingham PD, Marshall J, et al. Measurement of regional cerebral pH in human subjects using continuous inhalation of 11CO2 and positron emission tomography. J Cereb Blood Flow Metab. 1984;4:458–65.
Chareonthaitawee P, Kaufmann PA, Rimoldi O, Camici PG. Heterogeneity of resting and hyperemic myocardial blood flow in healthy humans. Cardiovasc Res. 2001;50:151–61.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
de Jong, H.W.A.M., Rijzewijk, L.J., Lubberink, M. et al. Kinetic models for analysing myocardial [11C]palmitate data. Eur J Nucl Med Mol Imaging 36, 966–978 (2009). https://doi.org/10.1007/s00259-008-1035-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-008-1035-3