Abstract
Objective
To evaluate if deep learning is a feasible approach for automated detection of supraspinatus tears on MRI.
Materials and methods
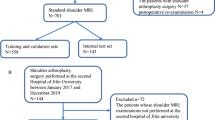
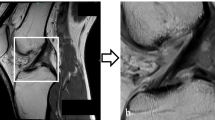
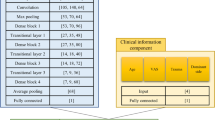
A total of 200 shoulder MRI studies performed between 2015 and 2019 were retrospectively obtained from our institutional database using a balanced random sampling of studies containing a full-thickness tear, partial-thickness tear, or intact supraspinatus tendon. A 3-stage pipeline was developed comprised of a slice selection network based on a pre-trained residual neural network (ResNet); a segmentation network based on an encoder-decoder network (U-Net); and a custom multi-input convolutional neural network (CNN) classifier. Binary reference labels were created following review of radiologist reports and images by a radiology fellow and consensus validation by two musculoskeletal radiologists. Twenty percent of the data was reserved as a holdout test set with the remaining 80% used for training and optimization under a fivefold cross-validation strategy. Classification and segmentation accuracy were evaluated using area under the receiver operating characteristic curve (AUROC) and Dice similarity coefficient, respectively. Baseline characteristics in correctly versus incorrectly classified cases were compared using independent sample t-test and chi-squared.
Results
Test sensitivity and specificity of the classifier at the optimal Youden’s index were 85.0% (95% CI: 62.1–96.8%) and 85.0% (95% CI: 62.1–96.8%), respectively. AUROC was 0.943 (95% CI: 0.820–0.991). Dice segmentation accuracy was 0.814 (95% CI: 0.805–0.826). There was no significant difference in AUROC between 1.5 T and 3.0 T studies. Sub-analysis showed superior sensitivity on full-thickness (100%) versus partial-thickness (72.5%) subgroups.
Data conclusion
Deep learning is a feasible approach to detect supraspinatus tears on MRI.







Similar content being viewed by others
References
Gyftopoulos S, Lin D, Knoll F, Doshi AM, Rodrigues TC, Recht MP. Artificial intelligence in musculoskeletal imaging: current status and future directions. AJR Am J Roentgenol. 2019;213:506–13.
Parker W, Forster BB. Artificial intelligence in sports medicine radiology: what’s coming? Br J Sports Med. 2018;53(19):1201–2.
Yamaguchi K, Ditsios K, Middleton WD, Hildebolt CF, Galatz LMTS. Morphological features of rotator cuff disease. A comparison of asymptomatic and symptomatic shoulders. J Bone Joint Surg Am. 2006;88(8):1699–704.
Yamamoto A, Takagishi K, Osawa T, Yanagawa T, Nakajima D, Shitara H, et al. Prevalence and risk factors of a rotator cuff tear in the general population. J Shoulder Elbow Surg. 2010;19:116–20.
Seida JC, LeBlanc C, Schouten JR, Mousavi SS, Hartling L, Vandermeer B, et al. Systematic review: nonoperative and operative treatments for rotator cuff tears. Ann Intern Med. 2010;2010:246–55.
Cofield RH, Parvizi J, Hoffmeyer PJ, Lanzer WL, Ilstrup DM, Rowland CM. Surgical repair of chronic rotator cuff tears: a prospective long-term study. J Bone Joint Surg Am. 2001;83:71–7.
Moosmayer S, Lund G, Seljom US, Haldorsen B, Svege IC, Hennig T, et al. At a 10-year follow-up, tendon repair is superior to physiotherapy in the treatment of small and medium-sized rotator cuff tears. J Bone Joint Surg Am. 2019;101:1050–60.
Tashjian RZ. Epidemiology, Natural history, and indications for treatment of rotator cuff tears. Clin Sports Med. 2012;31:589–604.
Roy J-S, Braën C, Leblond J, Desmeules F, Dionne CE, MacDermid JC, et al. Diagnostic accuracy of ultrasonography, MRI and MR arthrography in the characterisation of rotator cuff disorders: a systematic review and meta-analysis. Br J Sports Med. 2019;49:1316–28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25677796. (BMJ Publishing Group Ltd and British Association of Sport and Exercise Medicine)
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Meskó B, Görög M. A short guide for medical professionals in the era of artificial intelligence. npj Digit Med. 2020.
Kijowski R, Liu F, Caliva F, Pedoia V. Deep learning for lesion detection, progression, and prediction of musculoskeletal disease. J Magnet Reso. Imaging 2020:1607–19. Available from: https://pubmed.ncbi.nlm.nih.gov/31763739/ (John Wiley and Sons Inc)
Chea P, Mandell JC. Current applications and future directions of deep learning in musculoskeletal radiology. Skeletal Radiol. 2020:183–97. Available from: https://pubmed.ncbi.nlm.nih.gov/31377836/ (Springer)
Germann C, Marbach G, Civardi F, Fucentese SF, Fritz J, Sutter R, et al. Deep convolutional neural network-based diagnosis of anterior cruciate ligament tears: performance comparison of homogenous versus heterogeneous knee MRI cohorts with different pulse sequence protocols and 1.5-T and 3-T magnetic field strengths. Invest Radiol. 2020;55:499–506. Available from: https://pubmed.ncbi.nlm.nih.gov/32168039/ (Lippincott Williams and Wilkins)
Wang S, Su Z, Ying L, Peng X, Zhu S, Liang F, et al. Accelerating magnetic resonance imaging via deep learning. Proc - Int Symp Biomed Imaging. IEEE Comput Soc. 2016:514–7.
Medina G, Buckless CG, Thomasson E, Oh LS, Torriani M. Deep learning method for segmentation of rotator cuff muscles on MR images. Skeletal Radiol. 2021;50:683–92. Available from: https://pubmed.ncbi.nlm.nih.gov/32939590/ (Springer Science and Business Media Deutschland GmbH)
Wang G, Han Y. Convolutional neural network for automatically segmenting magnetic resonance images of the shoulder joint. Comput Methods Programs Biomed. 2020:200. Available from: https://pubmed.ncbi.nlm.nih.gov/33309302/ (Elsevier Ireland Ltd)
Conze PH, Brochard S, Burdin V, Sheehan FT, Pons C. Healthy versus pathological learning transferability in shoulder muscle MRI segmentation using deep convolutional encoder-decoders. Comput Med Imaging Graph. 2020;83:101733.
Kim JY, Ro K, You S, Nam BR, Yook S, Park HS, et al. Development of an automatic muscle atrophy measuring algorithm to calculate the ratio of supraspinatus in supraspinous fossa using deep learning. Comput Methods Programs Biomed. 2019;182:105063.
Cantarelli Rodrigues T, Deniz CM, Alaia EF, Gorelik N, Babb JS, Dublin J, et al. Three-dimensional MRI bone models of the glenohumeral joint using deep learning: evaluation of normal anatomy and glenoid bone loss. Radiol Artif. 2020;2:e190116. https://doi.org/10.1148/ryai.2020190116 (Radiological Society of North America (RSNA)).
Ro K, Kim JY, Park H, Cho BH, Kim IY, Shim SB, et al. Deep-learning framework and computer assisted fatty infiltration analysis for the supraspinatus muscle in MRI. Sci Rep. 2021;11:15065.
Shim E, Kim JY, Yoon JP, Ki SY, Lho T, Kim Y, et al. Automated rotator cuff tear classification using 3D convolutional neural network. Sci Rep. 2020;10:1–9. https://doi.org/10.1038/s41598-020-72357-0.
Lin CC, Wang CN, Ou YK, Fu J. Combined image enhancement, feature extraction, and classification protocol to improve detection and diagnosis of rotator-cuff tears on MR imaging. Jpn Soc Magnet Reson Med. 2014;13:155–66. Available from: https://pubmed.ncbi.nlm.nih.gov/24990467/
Reinus WR, Shady KL, Mirowitz SA, Totty WG. MR diagnosis of rotator cuff tears of the shoulder: value of using T2- weighted fat-saturated images. Am J Roentgenol . 1995;164:1451–5. Available from: https://pubmed.ncbi.nlm.nih.gov/7754891/
Patte D. Classification of rotator cuff lesions. Clin Orthop Relat Res. 1990. p. 81–6.
Ellman H. Diagnosis and treatment of incomplete rotator cuff tears. Clin Orthop Relat Res. 1990:64–74. Available from: https://europepmc.org/article/med/2182260
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. IEEE Comput Soc. 2016: 770–8. Available from: http://image-net.org/challenges/LSVRC/2015/
Dutta A, Zisserman A. The VIA annotation software for images, audio and video. MM 2019 - Proc 27th ACM Int Conf Multimed. New York, NY, USA: Association for Computing Machinery, Inc. 2019 :2276–9. https://doi.org/10.1145/3343031.3350535
Liu F, Zhou Z, Samsonov A, Blankenbaker D, Larison W, Kanarek A, et al. Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology. Radiological Society of North America Inc. 2018;289:160–9. https://doi.org/10.1148/radiol.2018172986
Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging. 2019
Roblot V, Giret Y, Bou Antoun M, Morillot C, Chassin X, Cotten A, et al. Artificial intelligence to diagnose meniscus tears on MRI. Diagn Interv Imaging [Internet]. Elsevier Masson SAS; 2019 [cited 2020 May 25];100:243–9. Available from: https://linkinghub.elsevier.com/retrieve/pii/S2211568419300555
Chang PD, Wong TT, Rasiej MJ. Deep learning for detection of complete anterior cruciate ligament tear. J Digit Imaging. 2019;32:980–6 (Elsevier Masson SAS).
Fritz B, Fritz J. Artificial intelligence for MRI diagnosis of joints: a scoping review of the current state-of-the-art of deep learning-based approaches. Skelet Radiol.2021:1–15. https://doi.org/10.1007/s00256-021-03830-8 (Springer)
Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. Saria S, editor. PLOS Med. 2018;15:e1002699 (Public Library of Science).
Kim MJ, Park H, Kim JY, Van Hoecke S, De Neve W. Towards diagnosis of rotator cuff tears in 3-D MRI using 3-D convolutional neural networks. Work Comput Biol Int Conf Mach Learn Proc. 2019. Available from: http://hdl.handle.net/1854/LU-8632543
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 3rd Int Conf Learn Represent ICLR. 2015. Available from: http://www.robots.ox.ac.uk/
Kim S, Lee D, Park S, Oh KS, Chung SW, Kim Y. Automatic segmentation of supraspinatus from MRI by internal shape fitting and autocorrection. Comput Methods Programs Biomed. 2017;140:165–74. Available from: https://pubmed.ncbi.nlm.nih.gov/28254072/ (Elsevier Ireland Ltd)
Razmjou H, Lincoln S, Macritchie I, Richards RR, Medeiros D, Elmaraghy A. Sex and gender disparity in pathology, disability, referral pattern, and wait time for surgery in workers with shoulder injury. BMC Musculoskelet Disord. 2016;17:401.
Ganal E, Ho CP, Wilson KJ, Surowiec RK, Smith WS, Dornan GJ, et al. Quantitative MRI characterization of arthroscopically verified supraspinatus pathology: comparison of tendon tears, tendinosis and asymptomatic supraspinatus tendons with T2 mapping. Knee Surg Sport Traumatol Arthrosc. 2016;24:2216–24. https://doi.org/10.1007/s00167-015-3547-2.
Schick F. Tissue segmentation: a crucial tool for quantitative MRI and visualization of anatomical structures. Magnet Reson Mater Physics Biol Med. 2016:89–93. Available from: https://pubmed.ncbi.nlm.nih.gov/27052370/ (Springer Verlag)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflict of interest
Author FJR declares that he is a Director of Medical Affairs for Imagia Cybernetics Inc.
Authors JY, LC, YN, PS, and AS have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yao, J., Chepelev, L., Nisha, Y. et al. Evaluation of a deep learning method for the automated detection of supraspinatus tears on MRI. Skeletal Radiol 51, 1765–1775 (2022). https://doi.org/10.1007/s00256-022-04008-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00256-022-04008-6