Abstract
A total no. of 65 Salmonella enterica isolates recovered from food samples, feces of diarrheic calves, poultry, and hospital patient in large five cities at Northern West Egypt were obtained from the Department of Microbiology, Faculty of Veterinary Medicine, Alexandria University, Alexandria, Egypt. The 65 Salmonella enterica isolates had the invA gene were grouped into 11 Salmonella enterica serovars with dominance of S. Enteritidis and S. Kentucky serovars. Their resistance pattern were characterized by using 18 antibiotics from different classes. Approximately 80% of the isolates were multidrug resistant (MDR). Enterobacterial repetitive intergenic consequences polymerase chain reaction (ERIC-PCR) typing of 7 strains of S. Enteritidis showed 5 clusters with dissimilarity 25%. S. Enteritidis clusters in 2 main groups A and B. Group A have 2 human strain (HE2 and HE3) and one food origin (FE7) with a similarity 99%. Group B divided into B1 (FE2) and B2 (FE3) with a similarity ratio ≥ 93%, while ERIC-PCR analysis of 5 strains of S. Kentucky revealed 4 ERIC types, clustered in 2 main groups A and B with similarity 75%. We studied the effect of silver nanoparticles (Ag-NPs) on 10 antibiotic resistant strains of S. Enteritidis and S. Kentucky. The broth microdilution minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were detected. Evaluation of the affection using scanning electron microscopy (SEM) and transmission electron microscopy (TEM) showed different ratios of Ag-NPs and microorganism as well as at different contact time ended finally with morphological alteration of the bacteria. We submitted new method in vivo to explore the activity of nanosilver in chicken.
Key points
• Importance of ERIC-PCR to determine the relatedness between Salmonella isolates.
• Effect of silver nanoparticles to confront the antibacterial resistance.
• Studying the effect of silver nanoparticles in vivo on infected chicken with Salmonella.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There are over 2500 serovars of Salmonella enterica (Andrews-Polymenis et al. 2010). Any serovar is thought to be capable of generating varied degrees of intestinal disease in people (Forshell and Wierup 2006). The majority of them are broad host range pathogens that infect a wide range of hosts, with only a few being host specific (Saroj et al. 2009); some serotypes are only found in certain parts of the world (Brands 2006), infecting animals, poultry, and fish, and are the leading cause of foodborne illness in humans globally (ECDC 2013).
According to the WHO, since 1990, Salmonella Enteritidis has been considered the most common cause of gastroenteritis worldwide (Chaitram et al. 2003) and salmonellosis in bovine, ovine, and poultry (Suh and Song 2006; Firoozeh et al. 2012; Dutta et al. 2012; Guizelini et al. 2019).
Meanwhile, Salmonella Kentucky isolation has been increased during recent years (Saroj et al. 2009; Osman et al. 2010a, b; Abd El-Ghany et al. 2012; Barua et al. 2012; Zahran et al. 2020) and as food borne pathogen both isolated by Shah et al. (2017) Shivaning Karabasanavar et al. (2020), Amin et al. (2021), Gawish et al. (2021), and Adel et al. (2021).
We require accurate subtyping information of strains to identify potential sources of infection, trace cross contamination, and pinpoint particularly virulent strains for efficient epidemiological surveillance and management of Salmonella species (Tenover et al. 1997; MacCannell 2013).
Increasing MDR S. enterica to cephalosporin and fluoroquinolones as critically important recommended treatment option (Chen and Schluesener 2008) will lead to increased severity, morbidity, and mortality of salmonellosis in humans and subsequently the use of the last line antimicrobials, e.g., cephapems (WHO 2021).
The prevalence of MDR S. enterica in Egypt, detected from retail meat samples, was 69.8% in 2010 and 82.4% and 100% in 2020 (Adel et al. 2021; Awad et al. 2020).
Remarkably, Egypt was formerly thought to be the source of highly drug-resistant S. Kentucky sequence type 198 (ST198-CipR) in Europe (Hawkey et al. 2019; Coipan et al. 2020), which was recently isolated from broilers in Lebanon (El Hage et al. 2020).
Several molecular techniques for typing Salmonellae have been proposed; the enterobacterial repetitive intergenic consequences polymerase chain reaction (ERIC–PCR), a simple technique of random amplified polymorphic DNA, has been successfully applied in genotyping of microbial pathogens, including gene mapping, detection strain diversity, population analysis, epidemiology, and demonstration of phylogenetic and taxonomic relationship (Li et al. 2009), without prior knowledge of target genome sequences (Maslow and Mulligan 1996; Stefańska et al. 2008; Li et al. 2009), faster with highest discriminatory power (Guimarães Ade et al. 2011) is an economical (Ranjbar et al. 2014) and capable of amplifying tiny amount of microbial DNA sequence.
As a result of the developed resistance of variant Salmonella species to antibiotics, which has become a major public health concern (Silver et al. 2006; Akinyemi et al. 2011), traditional antibiotics are being replaced by new alternative technologies such as nanotechnology, which has a wide range of potential applications in human and veterinary medicine (Rudramurthy et al. 2016). Silver nanoparticles are a suitable alternative among metallic nanoparticles with antibacterial activity because, in addition to possessing a strong antibacterial profile, they are also reasonably affordable to produce (Lee et al. 2007; Pal et al. 2007; Zhang et al. 2008; EL-sherif and Ali 2020) When comparing the studied nanoparticles, those with very low levels of MIC and MBC should be a focus in study, with concentration treatment and genus taken into consideration.
This work aimed to clarify the Salmonella enterica serovars and the benefit of silver nanoparticles (Ag-NPs) in the fight against the MDR bacterial strains. In the Northern West Egypt, starting from identification, antibiotic susceptibility testing, and the ability of silver nanoparticles in their application alone, take the opportunity of chicken as can be employed as a lab animal and host in vitro and in vivo. A specific attention to know if its antibacterial efficacy affected by methods of synthesis, concentration, time, and Salmonella serovars treated.
Material and Methods
Isolation and identification of Salmonella enterica from collected food samples
The preparation of all samples culturing and isolation of Salmonella was done according to the ISO 6579 (ISO 2002 and 2017). For isolation of Salmonella, swabs taken from humans or days old broiler chicks were performed as recorded by FDA.
Identification of presumed Salmonella spp. was carried out by morphological and cultural characteristics following standard microbiological methods (Washington winner et al. 2001; Quinn et al. 2013)
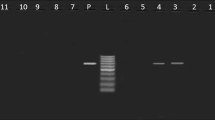
Typical Salmonella morphology samples identified biochemically, were confirmed by PCR for the presence of invA gene as shown in Fig. 1, further serotyped using specific Salmonella O and H antisera (Difco, Franklien lakes, NJ, USA). All the serological identification and molecular characterization were performed at the Animal Health Research Institute, Dokki, Giza, Egypt.
Agarose gel electrophoresis showing specific PCR using primer set for the invA gene. In this photo “Pos” stands for positive control, “Neg”; negative control and numbers indicate lanes with positive and negative isolates lane L (100–600 bp marker): all isolates (10) were positive that show specific band at 284 bp
Revival of bacterial strains
All bacteria isolated within 2020–2021 were revived in BHI broth (DIFICO) by overnight incubation at 37 °C, followed by plating on MacConkey’s agar (DIFICO) and confirmed serologically.
Antimicrobial susceptibility testing
For testing of antibiotic used, see Table 1; Kirby–Bauer disk diffusion assay was performed according to the standards and interpretive criteria described by clinical and laboratory standard institute (CLSI 2018).
The reference strain, Escherichia coli ATCC 25,922, was included as quality control strains showed resistance to antibiotics from at least three different classes considered as MDR — multidrug resistant (Magiorakos et al., 2012). The identity of S. Enteritidis was confirmed by repetitive sequence PCR using primers described the resistant Strain previously (Suh and Song 2006).
For MDR Salmonella Kentucky to be used for Ag-NP effect investigation, all the serological identification and molecular characterization were performed at the Animal Health Research Institute, Dokki, Giza, Egypt.
ERIC-PCR
Primer pairs for ERIC-PCR amplification were as follows: ERIC IR (5-ATGAACTCCTGGGGATTCAC-3) and ERIC-2 (5-AAGTAAGTACTGGGGTGAGCG C-3) amplification and condition were performed as usual (Versalovic et al. 1991). The size of the amplified fragments was determined after electrophoresis in a submerged agarose gel (1.5%) (Sambrook et al. 1989).
Ag-NP synthesis methods’
The different Ag-NPs that varied in size, synthesis method, and properties are summarized in Table 2. Silver nitrate Ag No3; 99.9%, Sigma Aldrich, st, l0 Mo, USA was used.
Susceptibility of Salmonella spp. to Ag-NPs
The broth microdilution briefly minimum inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) were conducted to measure the in vitro activity of Ag-NPs against each bacterial strain as conducted by (Wikler et al. 2008).
Ultra structure observations
Particle size and shape were determined by TEM (transmission electron microscope) and SEM (scanning electron microscope) utilization (Samberg et al. 2011); images were taken prior to post staining to show the location of Ag-NPs relative to bacteria and post stained with lead citrate, uranyl acetate in order to visualize cell morphology and membrane integrity.
In vivo evaluation of Ag-NP effect on S. Kentucky virulence
This in vivo evaluation Ag-NP effect of silver nanoparticles on the virulence of S. Kentucky was carried out by inoculating it in newly hatched days old broiler chicks from the Cobb lineage of breeding hens instead of SPF chicks. Days-old chicks were free from Salmonella infection (–v slide agglutination test and Salmonella isolation culture free).
The birds were kept in 32 °C heated environment and housed in isolated boxes with 30 cm length × 55 cm height × 35 cm breadth. Drinkable water and antibiotic free commercial food were provided.
The S. Kentucky was spread on XLD agar and incubated for 24 h at 37 °C. Three colonies were selected and removed to 10 ml of BHI broth and incubated for 24 h at 37 °C.
The chicks were randomized into three groups. Each group contains five chicks; the first group of chicks was infected by crop gavage with 0.5 ml of previously prepared broth culture containing 1 × 106 colony forming units of the S. Kentucky (Dhillon et al. 2001).
The second group was infected with S. Kentucky and 20 mg/ml silver nanoparticles, and was observed for 1 week and record observation (Osman et al. 2010a). The third group was injected with silver nanoparticles only.
All broiler chicks were investigated daily from the second day after injection for detecting shedding of Salmonella in feces by culturing on XLD medium.
At the end of the week, the chicks were euthanized through cervical dislocation to detect the presence of Salmonella spp. from re-isolation and slide agglutination reaction with somatic (O) antisera from liver and spleen (Borsoi et al. 2009).
The macroscopic lesions were observed such as airsacculitis, peritonitis, perihepatitis, pericarditis, and cellulitis, in the dead or killed chicks.
The chicks that survived until the seventh day to tenth day were killed by cervical dislocation, necropsied, and evaluated as explained before.
Results
A total of 65 Salmonella enterica strains included from 11 serovar species were revealed from different food samples including, chicken, beef, goat’s meat, hamburger, milk, cheese, and the feces of diarrheic calves and poultry. The investigation of 5 large cities in Northern West Egypt, Matrouh, Alexandria, Damnhour, Mahmoudia, and Desouq during the period time 2020–2021 ended by investigation of days old chick isolation of Salmonellae (n = 3 S. Kentucky, n = 3 S. London, one S. Ohio) at 2022 with the dominance of the serovar Salmonella Enteritidis and emergency of S. Kentucky among the most prevalence serotypes. All the types showed resistance to at least 3 of these antibiotics (ampicillin, oxacillin, streptomycin, and tetracyclin). Most of strains showed MDR phenotype (multidrug resistant) resist more than 3 classes of antibacterial as shown in Table 1.
ERIC-PCR typing of 7 strains S. Enteritidis showed 5 ERIC-PCR type clusters. The maximum dissimilarity was 25%, and these in a common band between all strains. S. Enteritidis clusters in 2 main groups A and B. Group A have 2 human strain HE2 and HE3 and one food origin FE7 with a similarity 99% in its subdivision A1 and A2 composed of one food origin (F1). Group B divided into B1 (FE2) only and B2 (FE3) only with a similarity ratio ≥ 93%. ERIC-PCR analysis of 5 strains S. Kentucky showed 4 ERIC-PCR types, clustered in 2 main groups A and B with similarity 75%. Group A composed of HK1, HK2 human origin, and FK2 from food. The subdivided A1 composed only from HK1 and HK2 with a similarity 99%, while A2 composed only from FK2. The similarity group between A1 and A2 was 93%. Group B composed of FK1 in subgroup B1 and FK5 in subgroup B2 with a similarity ≥ ratio 94% as shown in Figs. 2 and 3.
ERIC-PCR of Salmonella Enteritidis. A. ERIC-PCR finger printing of 7 S. Enteritidis isolates in 1.5% agarose gel, L: 100 bp molecular marker, HE1, HE2 and HE3: S. Enteritidis isolates from human origin, FE1, FE2, FE3 and FE7: S. Enteritidis isolates from food origin. B. Dendrogram showing the relatedness of 7 S. Enteritidis isolates using SPSS software program, HE1, HE2 and HE3: S. Enteritis isolates from human origin, FE1, FE2, FE3 and FE7: S. Enteritidis isolates from food origin
ERIC-PCR of S. Kentucky. A. ERIC-PCR finger printing of 5 S. Kentucky isolates in 1.5% agarose gel, L: 100bp molecular marker, FK1, FK2 and FK5: S. Kentucky isolates from food origin, HK1 and HK2: S. Kentucky isolates from human origin. B. Dendrogram showing the relatedness of 5 S. Kentucky isolates using SPSS software program, FK1, FK2 and FK5: S. Kentucky isolates from food origin, HK1 and HK2: S. Kentucky isolates from human origin
Results of Ag-NP effect (50 ppm) on S. Enteritidis cells revealed by SEM image after treatment for 3 h and 24 h
The results of Ag-NP effects on S. Enteritidis bacterial cells revealed by SEM, S. Enteritidis cells with Ag-NP particles at concentration of 50 ppm after 3 h, we can notice morphological damage disruption of the cell wall, and can noted S. Enteritidis cells with Ag-NP particles at concentration of 50 ppm after 24 h Ag-NPs; the cell and complete bacterial lysis was observed at 24 h as shown in Fig. 6.
A Untreated cells S. Enteritidis showing intact cells. Cells had uniform electron density, straight with rounded end. B Treated S. Enteritidis cells with Ag-NP particles at concentration of 50 ppm after 3 h by SEM; we can notice morphological damage–disruption of cell wall. C S. Enteritidis cells after 24 h treatment with 50 ppm Ag NPs presented rupture in the cell wall and also losing shape of bacterial cell and complete bacterial lysis only some bacteria loose the rounded ends, the ends much as pointed and less extent in its width
The results of Ag-NP effects (50 ppm) on S. Kentucky cells revealed by SEM image, after treatment for 3 h and 24 h
The results of Ag-NP effect on S. Kentucky bacterial cells revealed by SEM, S. Kentucky cells with Ag-NP particles at concentration of 50 ppm after 3 h, we can notice morphological image disruption of the cell wall and can noted S. Kentucky cells with Ag-NP particles at concentration of 50 ppm after 24 h, the cells presented rupture in the cell wall, and also, losing shape of bacterial cell and complete bacterial lysis was observed at 24 h as shown in Fig. 7.
A Untreated cells S. Kentucky showing intact cells. Cells had uniform electron density, straight with rounded end. B Treated S. Kentucky cells with Ag-NP particles at concentration of 50 ppm after 3 h by SEM, we can notice morphological damage–disruption of cell wall, C S. Kentucky cells after 24 h treatment with 50 ppm Ag-NPs presented rupture in the cell wall and also, losing shape of bacterial cell and complete bacterial lysis. Some bacteria decreased in width and other are completely damaged, losing its rounded ends
The preparing of silver nanoparticles (Ag-NPs) showed its characterization by UV–VIS spectroscopy, transmission electron microscope (TEM), and scanning electron microscope (SEM) average size 26.5 nm, 30–40 nm, and 45 ± 5 nm have the detected (MIC) minimum inhibitory concentration showed strong antibacterial activity. The results of disc diffusion method showed no significant difference due to different sizes, but the time of contact and concentration of Ag-NPs directly affected the bacterium as from the electron microscopy images.
We noticed the MIC and MBC of S. London half that of S. Kentucky as in previous study the same strain using Zn-NPs obtained by (Wang et al. 2018) method. Interestingly, MIC and MBC were also the same; i.e., MIC was 2.5 µg/ml and 5 µg/ml for S. London and S. Kentucky, respectively, and MBC was 5 µg/ml and 10 µg/ml for S. London and S. Kentucky, respectively.
SEM images show all the untreated control cells have intact and smooth surface. Both Salmonella spp. remained normally rod shaped, Ag-NPs adhered mainly to cell wall of S. Enteritidis, and some cells show penetration of Ag-NPs inside, thus modified the cell and characterized by the formation of “pits” allowed entry of Ag-NPs into the cell and may be death. At the site of Ag-NP adsorbed aggregation, there is widening of the periplasmic space in which Ag-NPs had accumulated the disrupted lysis cell membrane, damaged cytoplasm, and cell deformity.
In case of S. Kentucky Ag-NPs were able to damage the cell wall, but did not enter the cell, the cells appear to be shorter and more compact, suggesting there could be some leakage of the cellular content caused by the treatment; no lysis was noticed.
In vivo
Two out of five chicks injected only S. Kentucky died before the end of the first week, i.e., mortality rate 2/5. This group showed re-isolation of Salmonella on XLD agar from positive slide agglutination test and from liver and spleen. The pathological lesion was severe hepatitis with multiple pale foci and pericarditis, peritonitis, airsacculitis, and pneumonia.
Discussion
This is the primary nationwide survey for prevalence of Salmonella enterica serovar species isolated from calves, food, poultry, and human food in northern west provincial five cities Matrouh, Alexandria, Damanhour, Mahmoudia, and Desouq.
The serovar analysis has shown S. Enteritidis serovar as the dominant serovar and high-frequency S. Kentucky isolation. Recent reports from Brazil, Poland, Malaysia, China, and Greece have shown the same S. Enteritidis serovar with frequencies ranging from 34 to 86% which reveals coincident rise of the serovar around the world (Wen et al. 2017; Fernandes et al. 2006; Sadkowska-Todys and Czarkowski 2014; Spiliopoulou et al. 2007) and the high frequency of S. Kentucky was reported in Egypt (Osman et However, Salmonella species dominant serotypes have changed over time in various geographical locations (Fardsanei et al. 2016).
In antibiotic sensitivity testing approximately 3 from 4 strains of S. enterica were multidrug-resistant MDR, in which bacterial strains are resistant to more than three classes of antibiotics (Magiorakos et al. 2012), this was expected due to worldwide spreading of MDR bacteria (Hawkey et al. 2019; Coipan et al. 2020 and WHO 2021). As previously documented for clinical isolates, the variation between the five cities is related to reduce prescribing of particular drugs in certain cities (Gatto et al. 2006; Davin-Regli et al. 2008). The isolation of one strain of bacteria resistant to more than ten antibiotics in poultry has led us to fear that there will be no effective medicines available to treat resistant infections one day (Handayani et al., 2017). Chloramphenicol, which was blindly supplied by veterinarians in Egypt for diarrhoea, was found to stimulate the MDR response by activating the production of certain regulatory mRNA or other genes, according to Davin-Regli et al. (2008). While point mutations in DNA gyrase genes or activation of the efflux pump may cause enhanced resistance in food-borne isolates (Meakins et al. 2008), horizontal transfer and clonal expansion of resistance genes may occur among food-producing animals and humans (Hawkey 2008). The majority of developed countries utilize a regular surveillance and monitoring a system for antimicrobial drug resistance (AMP) that updated regularly to identify changes in antimicrobial resistance (WHO 2021), for example, national antimicrobial resistance monitoring system (NARMS) in the united states and the Danish integrated antimicrobial resistance monitoring and research program (DANMAP) in Denmark. The developing countries like Egypt did not have these systems (Vernet et al. 2014). Antibiotic resistance and serotyping methods (historically) were provided data to be used for short epidemiological studies, trends in well-defined geographical areas, and comparing between different countries (Tenover et al. 1997; Maccanell et al. 2013). Today, these methods in the increased MDR bacteria and time cost in my opinion do not have practical value as previously mentioned by Ranjbar et al. (2014). Recently, DNA-based typing method like ERIC-PCR subtype becomes indispensable to study the epidemiology of most microbial pathogens (Ranjbar et al. 2014). Our investigation used ERIC-PCR infer transmission of Salmonella Enteritidis from Mahmoudia city to Alexandria city as food born pathogen and cross transmission between calf and poultry in the same farm yards. Moreover, we reported 5 ERIC-PCR types of S. Enteritidis isolated from food and human. A previous study reported six ERIC types of S. Enteritidis isolated from food and patients from north Morocco (Ammari et al. 2009) and from southern Brazil revealed 3 ERIC types (Oliveira et al. 2007) and from India from diverse origin were categorized into clusters (Anjay et al. 2015) and in Iran into 4 clusters (Fardsanei et al. 2016) showed five different banding patterns with two major common types representing 76.6% of the 30 isolates they examined, each of which considered of both clinical and food isolates. The CT 3 only includes clinical, while CT4 included food samples.
For S. Kentucky, the 5 isolates (2 from human and 3 from food) were clustered in two main groups A and B subdivided into (AI, A and B1, B) giving 4 ERIC-PCR types with similarity 75%. The two human samples clustered in A1 with one food origin had 99% similarity, while that from food are in cluster A2. The similarity between A1 and A2 was 93%. B1 and B2 each composed of one from food origin with similarity ≥ 94%. ERIC-PCR results infer that all isolates which were phenotypically homogenous also genotypically homogenous were clonally dispersed among food, human population, animal, and poultry, and may continue to exist over considerable period of time on northern west Egypt and spread in different time occasion, supported the notion that infected animals, poultry, and humans are important source of contamination on the environment and food chain.
In vitro, the concentration of Ag-NPs and contact time directly affect the bacterial activity. As the increased of reactivity with decreasing particle size increased the number of attached cells due to large surface area, it provides better contact with microorganisms. The number in the different methods was used for preparing Ag-NP particles (Morones et al. 2005; Lok et al. 2006; Dror-Ehre et al. 2009; Kourmouli et al. 2018). Due to large surface area provide Ag-NPs, better contact with microorganism independent then size (Toker et al. 2013). The difference in method of synthesis provides different electrical charges that increase in the positive, less in neutral, and weak in negative (Abbaszadegan et al. 2015). This study showed that Ag-NPs of the exact same size and same synthesis method unyield vastly different MIC and MBC value simply on Salmonella enterica serovar species; time for interaction plays important role in damage and lysis in bacteria and initial concentration. The concentration and time of contact significantly affect the bacterial response. Kourmouli et al. (2018) concluded that the apparent antibacterial behavior is attributed to the ion Ag-NP release rather than to their unique size-dependent properties. Previously, the number of C.F.U. of Salmonella spp. was significantly reduced with increasing concentration (Raffi et al. 2008; Guzman et al. 2012; El- Sherif and Ali 2020) and the destruction and damage of cell wall lysis, expulsion of cellular content, mechanisms such as negative regulation of porins, chromosomal resistance genes, or plasmid with resistant genes have been proposed (Salas-Orozco et al. 2019). It has been implied by many authors that Ag-NPs are capable of attaching to bacterial cell membrane and as well as entry into cells (Pal et al. 2007; Dror- Ehre et al. 2009); they did not use the electron microscopy. Others reported that only Ag-NPs with diameter less than 10 nm were capable of entry E. coli and Pseudomonas aeruginosa (Morones et al. 2005), while 80 nm size can accumulate within after the addition of chloramphenicol (Xu et al. 2004). However, Samberg et al. (2011) showed ruptured and damaged bacteria with Ag-NP agglomerate nearby, but the electron microscopy image confirmed the actual penetration of Ag-NPs into whole bacteria. The success of Ag-NPs as effective antimicrobial is strongly strain dependent, since sensitivity to action of Ag-NPs, and MIC, thus is probably due to cell wall thickness differences. Berton et al. (2014), He et al. (2016), and Stoyanova et al. (2016) said that thus due to sensitivity, while Silver et al. (2006) is probably genetic factors specifically intrinsic of each strain including the presence of specific determinant of resistance, the possible mechanism of action is that the metal nanoparticles are carrying the positive charge and microbes the negative charges which create the electromagnetic attraction and microbe get oxidized (Rezaei Zarchi et al., 2010) or nanoparticles which react with thiol group (-SH) of the protein present in the cell surface of bacteria leads to lysis (Zhang 2013).
In vivo, in this work, silver nanoparticles (Ag-NPs) proved as proficient prevention and therapeutic agents due to their outstanding, physical mode of action (Meena et al. 2018). Many theories had been proposed the unclear blurred mechanism of action of Ag-NPs by which it exerts their antimicrobial effect, but two main hypotheses have been exposed: a direct interaction after adhesion on bacterium cell wall and the release of ionic silver (Gugala et al. 2022). Recently, reduction in the silver ions (Ag) concentration of polymer-coated Ag-NPs did not affect their antibacterial efficacy (Ashmore et al. 2018); mechanisms such as negative regulation porins, chromosomal resistance genes, or plasmid with resistance genes have been proposed (Salas-Orozco et al. 2019). Finally, in the twentieth century, a popular belief was that except for causing Angria, silver was relatively non-toxic to mammalian cells; however, studies conducted recently have shown that at the nanoscale, silver-based materials can exhibit significant toxicity to animals and human cells; these issues must be addressed before people rush to indulge into the nanosilver boom (Chen and Schluesener 2008). So, the silver nanoparticles could be used in the treatment in the intestinal tract of poultry (usual place of habitat and propagation of Salmonella), in this original research, a virulent MDR strain of S. Enteritidis fails to cause and continue its pathogenesis thus occur through combine Ag-NPs to S. Enteritidis cells in the intestinal tract of days old chicks broilers. Nokhodchi et al. (2012) in their review article clarify the challenge in drug delivery to combat Salmonella spp. and fail of new antibiotic to eradicate the pathogens completely, due to difficulty of transport of antibiotic retail through membrane (Drulis-Kawa and Dorotkiewicz-Jach 2010); reduced cell membrane permeability has been dedicated as a key mechanism of resistance to antibiotic (Davin-Regli et al. 2008), while nanoparticles adhere to cell membrane of Salmonella and in some species of Salmonella can release into the interior of the bacteria as in case of Salmonella Enteritidis, thus can interfere with bacterial resistance and infection mechanism which involve low membrane permeability or efflux system (Mugabe et al. 2006). But the dosage of silver nanoparticles differs according to its concentration (Ranjan et al. 2009). Generally, in the twentieth century, a popular belief was that except for causing Angria, silver was relatively nontoxic to mammalian cells. However, studies conducted in recent decades have shown that at the nanoscale, silver-based materials can exhibit significant toxicity to animal and human cells. These issues must be addressed before people rush to indulge into the nanosilver boom (Chen and Schluesener 2008); nowadays, nanoparticles have been used in disinfection textile fabrics, water disinfection, medicine and food packing, and preservation (García-Barrasa et al. 2011; Toker et al. 2013; Antonio et al. 2014; Mihindukulasuriya and lim 2014). We here report the results of the primary investigation for the prevalence of Salmonella enterica serovar spp. isolated from food, animals, poultry, and hospital patient in large five cities at Northern West Egypt. Results showed sources importance as vehicles for the dissemination of the Salmonella and posed a critical health risk for the populations. Electron microscopy effect on S. Enteritidis and S. Kentucky and antibiotic-resistant patterns of the isolated Salmonella were explored; therefore, close surveillance of antimicrobial resistance in bacteria should be established as a priority. Our data provide a base for further investigations.
Data availability
The datasets used and/or analyzed during the current study are available from corresponding author on reasonable request.
References
Abbaszadegan A, Ghahramani Y, Gholami A, Hemmateenejad B, Dorostkar S, Nabavizadeh M & Sharghi H (2015) The effect of charge at the surface of silver nanoparticles on antimicrobial activity against gram-positive and gram-negative bacteria: a preliminary study. J Nanomater 2015. https://doi.org/10.1155/2015/720654
Abd El-Ghany WA, El-Shafii SS, Hatem M (2012) A survey on Salmonella species isolated from chicken flocks in Egypt. AJAVA 7:489–501. https://doi.org/10.3923/ajava.2012.489.501
Abdelsalam NR, Fouda MM, Abdel-Megeed A, Ajarem J, Allam AA & El-Naggar ME (2019) Assessment of silver nanoparticles decorated starch and commercial zinc nanoparticles with respect to their genotoxicity on onion. Int J Biol Macromol, 133, 1008–1018. https://doi.org/10.1016/j.ijbiomac.2019.04.134s
Adel WA, Ahmed AM, Hegazy Y, Torky HA, Shimamoto T (2021) High prevalence of ESBL and plasmid-mediated quinolone resistance genes in Salmonella enterica isolated from retail meats and slaughterhouses in Egypt. Antibiotics 10:881. https://doi.org/10.3390/antibiotics10070881
Akinyemi KO, Iwalokun BA, Foli F, Oshodi K, Coker AO (2011) Prevalence of multiple drug resistance and screening of enterotoxin (stn) gene in Salmonella enterica serovars from water sources in Lagos, Nigeria. Public Health 125:65–71. https://doi.org/10.1016/j.puhe.2010.11.010
Amin HS, Torky HA, Ebied SK (2021) A contribution to characterization of Salmonella Serovars recovered from retail food and human being in Alexandria City. Alex J Vet Sci 71:19–28. https://doi.org/10.5455/ajvs.125368
Ammari S, Laglaoui A, En-Nanei L, Bertrand S, Wildemauwe C, Abid M (2009) Characterization of Salmonella Enteritidis isolated from foods and patients in northern Morocco. J Infect Dev Ctries 3:695–703. https://doi.org/10.3855/jidc.617
Andrews-Polymenis HL, BäUmler AJ, Mccormick BA, Fang FC (2010) Taming the elephant Salmonella biology, pathogenesis and prevention. Infect immun 78:2356–2369. https://doi.org/10.1128/2FIAI.00096-10
Anjay AK, Agarwal R, Ramees T, Dubal Z, Kaushik P, Kumar MS, Dudhe N, Milton A, Abhishek B, Shagufta B (2015) Molecular typing of Salmonella Typhimurium and S Enteritidis serovars from diverse origin by ERIC-PCR. J Pure Appl Microbiol 9:2627–2634. https://doi.org/10.1128/2FJCM.41.9.4388-4394.2003
Antonio JR, Antônio CR, Cardeal ILS, Ballavenuto JMA, Oliveira JR (2014) Nanotechnology in Dermatology an Bras Dermato 89:126–136. https://doi.org/10.1590/abd1806-4841.20142228
Ashmore D, Chaudhari A, Barlow B, Barlow B, Harper T, Vig K, Miller M, Singh S, Nelson E & Pillai S (2018) Evaluation of E. coli inhibition by plain and polymer-coated silver nanoparticles. Rev Inst Med Trop Sao Paulo, 60, e18. https://doi.org/10.1590/s1678-9946201860018
Awad A, Gwida M, Khalifa E, Sadat A (2020) Phenotypes, antibacterial-resistant profile, and virulence-associated genes of Salmonella serovars isolated from retail chicken meat in Egypt. Vet World 13:440–445. https://doi.org/10.1590/s1678-9946201860018
Barua H, Biswas PK, Olsen KE, Christensen JP (2012) Prevalence and characterization of motile Salmonella in commercial layer poultry farms in Bangladesh. PLoS ONE 7:e35914. https://doi.org/10.1371/journal.pone.0035914
Berton V, Montesi F, Losasso C, Facco D, Toffan A, Terregino C (2014) Study of the interaction between silver nanoparticles and Salmonella as revealed by transmission electron microscopy. J Prob Health. https://doi.org/10.4172/2329-8901.1000123
Borsoi A, Santin E, Santos L, Salle C, Moraes H, Nascimento V (2009) Inoculation of newly hatched broiler chicks with two Brazilian isolates of Salmonella Heidelberg strains with different virulence gene profiles, antimicrobial resistance, and pulsed field gel electrophoresis patterns to intestinal changes evaluation. Poult Sci 88:750–758. https://doi.org/10.3382/ps.2008-00466
Brands DA (2006) Deadly diseases and epidemics Salmonella, Chelsea House publisher, asubsidiary of heights cross communications, P. 102.
Chaitram JM, Jevitt LA, Lary S, Tenover FC (2003) The World Health Organization’s External Quality Assurance System Proficiency Testing Program has improved the accuracy of antimicrobial susceptibility testing and reporting among participating laboratories using NCCLS methods. J Clin Microbiol 41:2372–2377. https://doi.org/10.1128/JCM.41.6.2372-2377.2003
Chen X, Schluesener HJ (2008) Nanosilver: a nanoproduct in medical application. Toxicol Lett 176:1–12. https://doi.org/10.1016/j.toxlet.2007.10.004
CLSI (2018) Clinical and Laboratory Standards Institute performance standards for antimicrobial disk susceptibility tests, 26th ed.; Twenty third Informational Supplement M100 -S23. CLSI, Wayne
Coipan CE, Westrell T, Van Hoek A, Alm E, Kotila S, Berbers B, De Keersmaecker SCJ, Ceyssens PJ, Borg ML, Chattaway M, Mccormick J, Dallman TJ, Franz E (2020) Genomic epidemiology of emerging ESBL-producing Salmonella Kentucky bla (CTX-M-14b) in Europe. Emerg Microbes Infect 9(2124):2135. https://doi.org/10.1080/2F22221751.2020.1821582
DANMAP Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark https://backend.orbit.dtu.dk/ws/files/161713656/Rapport_DANMAP_2017.pdf [ accessed 13 july 2021]
Davin-Regli A, Bolla JM, James CE, Lavigne JP, Chevalier J, Garnotel E, Molitor A, Pagès JM (2008) Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr Drug Targets 9:750–759. https://doi.org/10.2174/138945008785747824
Dhillon A, Shivaprasad H, Roy P, Alisantosa B, Schaberg D, Bandli D, Johnson S (2001) Pathogenicity of environmental origin Salmonellas in specific pathogen-free chicks. Poult Sci 80:1323–1328. https://doi.org/10.1093/ps/80.9.1323
Dror-Ehre A, Mamane H, Belenkova T, Markovich G, Adin A (2009) Silver nanoparticle–E. coli colloidal interaction in water and effect on E. coli survival. J Colloid Interface Sci 339:521–526. https://doi.org/10.1016/j.jcis.2009.07.052
Drulis-Kawa Z, Dorotkiewicz-Jach A (2010) Liposomes as delivery systems for antibiotics. Int J Pharm 387:187–198. https://doi.org/10.1016/j.ijpharm.2009.11.033
Dutta T, Roychoudhury P, Rajkhowa T, Chandra R (2012) Characterization of Salmonella Enteritidis from two separate outbreaks of acute enteritis in Japanese quail (Coturnix coturnix japonica) and turkey (Melleagris gallopavo). I J S R 2:14–16. https://doi.org/10.15373/22778179/FEB2013/6
ECDC (2013) European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/en/search? S= Salmonella.
El Hage R, Losasso C, Longo A, Petrin S, Ricci A, Mathieu F, Abi Khattar Z, El Rayess Y (2020) Whole-genome characterisation of TEM-1 and CMY-2 β-lactamase-producing Salmonella Kentucky ST198 in Lebanese broiler chain. J Glob Antimicrob Resist 23:408–416. https://doi.org/10.1016/j.jgar.2020.11.002
Elsherif WM & DN Ali (2020) Antibacterial effect of silver nanoparticles on antibiotic resistant E. coli O157:H7 isolated from some dairy products. Bulg J Vet Med, 23, No 4, 432−442. https://doi.org/10.15547/bjvm.2019-0027
Fardsanei F, Nikkhahi F, Bakhshi B, Salehi TZ, Tamai IA, Soltan Dallal MM (2016) Molecular characterization of Salmonella enterica serotype Enteritidis isolates from food and human samples by serotyping, antimicrobial resistance, plasmid profiling, (GTG)5-PCR and ERIC-PCR. New Microbes New Infect 14:24–30. https://doi.org/10.1016/j.nmni.2016.07.016
Fernandes SA, Tavechio AT, Ghilardi ÂC, Dias ÂM, De Almeida IA, De Melo LC (2006) Salmonella serovars isolated from humans in São Paulo State, Brazil, 1996–2003. Rev Inst Med Trop S Paulo 48:179–184. https://doi.org/10.1590/S0036-46652006000400001
Firoozeh F, Zahraei-Salehi T, Shahcheraghi F, Karimi V, Aslani MM (2012) Characterization of class I integrons among Salmonella enterica serovar Enteritidis isolated from humans and poultry. FEMS Immunol Med Microbiol 64:237–243. https://doi.org/10.1111/j.1574-695X.2011.00883.x
Forshell, LP & Wierup, M (2006) Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech, 25, 541–54. https://www.researchgate.net/publication/6702103_Salmonella_contamination_A_significant_challenge_to_the_global_marketing_of_animal_food_products
García-Barrasa J, López-De-Luzuriaga J, Monge M (2011) Silver nanoparticles: synthesis through chemical methods in solution and biomedical applications. Cent Eur J Chem 9:7–19. https://doi.org/10.2478/s11532-010-0124-x
Gatto AJ, Peters TM, Green J, Fisher IS, Gill ON, O’brien SJ, Maguire C, Berghold C, Lederer I, Gerner-Smidt P, Torpdah M, Siitonen A, Lukinmaa S, Tschäpe H, Prager R, Luzzi I, Dionisi AM, Wk VDZ, Heck M, Coia J, Brown D, Usera M, Echeita A, Threlfall EJ (2006) Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S Typhimurium definitive phage type 104 in nine European countries 2000-2004, results of an international multi-centre study. Epidemiol Infect 134(729):36. https://doi.org/10.1017/S0950268805005820
Gawish MF, Ahmed AM, Torky HA, Shimamoto T (2021) Prevalence of extended-spectrum β-lactamase (ESBL)-producing Salmonella enterica from retail fishes in Egypt: A major threat to public health. Int J Food Microbiol 351:109268. https://doi.org/10.1016/j.ijfoodmicro.2021.109268
Gugala N, Salazar-Alemán DA, Chua G & Turner RJ (2022) Using a chemical genetic screen to enhance our understanding of the antimicrobial properties of copper. Metallomics, 14. https://doi.org/10.1093/mtomcs/mfab071
Guimarães Ade S, Dorneles EM, Andrade GI, Lage AP, Miyoshi A, Azevedo V, Gouveia AM, Heinemann MB (2011) Molecular characterization of Corynebacterium pseudotuberculosis isolates using ERIC-PCR. Vet Microbiol 153:299–306. https://doi.org/10.1016/j.vetmic.2011.06.002
Guizelini CC, Pupin RC, Leal CR, Ramos CA, Pavarini SP, Gomes DC, Martins TB, Lemos RA (2019) Salmonellosis in calves without intestinal lesions. Pesq Vet Bras 39:580–586. https://doi.org/10.1590/1678-5150-PVB-6328
Guzman M, Dille J, Godet S (2012) Synthesis and antibacterial activity of silver nanoparticles against gram-positive and gram-negative bacteria. Nanomedicine 8:37–45. https://doi.org/10.1016/j.nano.2011.05.007
Handayani RS, Siahaan S & Herman MJ (2017) Antimicrobial resistance and its control policy implementation in hospital in Indonesia. Journal Penelitian dan Pengembangan Pelayanan Kesehatan, 131–140. https://doi.org/10.22435/jpppk.v1i2.537
Hasan T & Lafta I (2021) Identification and antimicrobial susceptibility profiles of Salmonella spp. isolated from chicken flocks and their feed and water in Karbala, Iraq. Indian J Ecol. https://www.researchgate.net/publication/354761731
Hawkey J, Le Hello S, Doublet B, Granier SA, Hendriksen RS, Fricke WF, Ceyssens PJ, Gomart C, Billman-Jacobe H, Holt KE & Weill FX (2019) Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb Genom, 5. https://doi.org/10.1099/mgen.0.000269
Hawkey P (2008) Molecular epidemiology of clinically significant antibiotic resistance genes. Br J Pharmacol 153:S406–S413. https://doi.org/10.1038/sj.bjp.0707632
He Y, Ingudam S, Reed S, Gehring A, Strobaugh TP, Irwin P (2016) Study on the mechanism of antibacterial action of magnesium oxide nanoparticles against foodborne pathogens. J Nanobiotechnology 14:54. https://doi.org/10.1186/s12951-016-0202-0
ISO (2002) International Organization for Standardization. 6579:2002. Microbiology of food and animal feeding stuffs. Horizontal method for the detection of Salmonella Spp. International Organization for Standardization, Geneva, Switzerland.
ISO6579–1 (2017) Microbiology of the food chain — horizontal method for the detection, enumeration and serotyping of Salmonella — part 1. Detection of Salmonella Spp. International Standards for Organization, Geneva, Switzerland.
Kourmouli A, Valenti M, Van Rijn E, Beaumont H J, Kalantzi OI, Schmidt-Ott A & Biskos G (2018) Can disc diffusion susceptibility tests assess the antimicrobial activity of engineered nanoparticles? J Nanoparticle Res., 20, 1–6. https://doi.org/10.1007/s11051-018-4152-3
Lee HY, Park HK, Lee YM, Kim K, Park SB (2007) A practical procedure for producing silver nanocoated fabric and its antibacterial evaluation for biomedical applications. Chem Commun :2959-2961. https://doi.org/10.1039/B703034G
Li W, Raoult D, Fournier PE (2009) Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33:892–916. https://doi.org/10.1111/j.1574-6976.2009.00182.x
Lok CN, Ho CM, Chen R, He QY, Yu WY, Sun H, Tam PKH, Chiu JF, Che CM (2006) Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J Proteome Res 5:916–924. https://doi.org/10.1021/pr0504079
Maccannell D (2013) Bacterial strain typing. Clin Lab Med 33:629–650. https://doi.org/10.1016/j.cll.2013.03.005
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Maslow J, Mulligan ME (1996) Epidemiologic typing systems. Infect Control Hosp Epidemiol 17:595–604. https://doi.org/10.1086/647395
Meakins S, Fisher IS, Berghold C, Gerner-Smidt P, Tschäpe H, Cormican M, Luzzi I, Schneider F, Wannett W, Coia J (2008) Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000–2004: a report from the Enter-net International Surveillance Network. Microb Drug Resist 14:31–35. https://doi.org/10.1089/mdr.2008.0777
Meena N, Sahni YP, Thakur DK, Singh R (2018) Applications of nanotechnology in veterinary therapeutics. J Entomol Zool Stud 6:167–175
Mihindukulasuriya SDF, Lim LT (2014) Nanotechnology development in food packaging A review. Trends Food Sci Technol 40(2):149167. https://doi.org/10.1016/j.tifs.2014.09.009
Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramírez JT, Yacaman MJ (2005) The bactericidal effect of silver nanoparticles. Nanotechnology 16:2346. https://doi.org/10.1088/0957-4484/16/10/059
Mugabe C, Halwani M, Azghani AO, Lafrenie RM, Omri A (2006) Mechanism of enhanced activity of liposome-entrapped aminoglycosides against resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:2016–2022. https://doi.org/10.1128/aac.01547-05
NARMS Antibiotic resistance threats in the united states 2019. Available online: https://www.cdc.gov/narms/index.html [ accessed in july 2021]
Nokhodchi A, Ghafourian T, & Mohammadi G (2012) Nanotechnology tools for efficient antibacterial delivery to Salmonella (pp. 139–168). Intech publishing group. https://doi.org/10.5772/31045
Oliveira SDD, Bessa MC, Santos LRD, Cardoso MRDI, Brandelli A, Canal CW (2007) Phenotypic and genotypic characterization of Salmonella Enteritidis isolates. Braz J Microbiol 38:720–728. https://doi.org/10.1590/S1517-83822007000400025
Osman KM, Moussa IM, Yousef AM, Aly MM, Radwan MI & Alwathnani H (2010a) Pathogenicity of some avian Salmonella serovars in two different animal models: SPF chickens and BALB/c mice. Environ We Int J Sci Tech 5:65–78. https://www.researchgate.net/publication/228519150_Pathogenicity_of_some_avian_Salmonella_serovars_in_two_different_animal_models_SPF_chickens_and_BALBc_mice
Osman KM, Yousef AM, Aly MM, Radwan MI (2010b) Salmonella spp. infection in imported 1-day-old chicks, ducklings, and turkey poults: a public health risk. Foodborne Pathog Dis., 7, 383–390. https://doi.org/10.1089/fpd.2009.0358
Pal S, Tak YK & Song JM (2007) Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl Environ Microbiol 73:1712–1720. https://doi.org/10.1128/aem.02218-06
Quinn PJ, Carter ME, Markey BK, Carter GR (2013) Clinical veterinary microbiology. Mosby, Missouri, USA
Raffi M, Hussain F, Bhatti T, Akhter J, Hameed A, Hasan M (2008) Antibacterial characterization of silver nanoparticles against E. coli ATCC-15224. J Mater Sci Technol 24:192–196. https://www.jmst.org/EN/Y2008/V24/I02/192
Ranjan A, Pothayee N, Seleem MN, Tyler RD Jr, Brenseke B, Sriranganathan N, Riffle JS, Kasimanickam R (2009) Antibacterial efficacy of core-shell nanostructures encapsulating gentamicin against an in vivo intracellular Salmonella model. Int J Nanomed 4:289–297. https://doi.org/10.2147/ijn.s7137
Ranjbar R, Karami A, Farshad S, Giammanco GM, Mammina C (2014) Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol 37:1–15. https://www.researchgate.net/publication/260218307_Typing_methods_used_in_the_molecular_epidemiology_of_microbial_pathogens_a_how-to_guide
Rezaei Zarchi S, Javed A, Ghani MJ, Soufian S, Barzegari Firouzabadi F, Bayandori Moghadam AM & Mirjalili SH (2010) Comparative study of antimicrobial activities of TiO2 and CdO nanoparticles against the pathogenic strain of Escherichia coli. IJP 5(2):83-89. https://www.sid.ir/en/journal/ViewPaper.aspx?id=169005
Rudramurthy GR, Swamy MK, Sinniah UR, Ghasemzadeh A (2016) Nanoparticles: alternatives against drug-resistant pathogenic microbes. Molecules 21. https://doi.org/10.3390/molecules21070836
Sadkowska-Todys M, Czarkowski MP (2014) Salmonellosis in Poland in 2012. Przegl Epidemiol 68(243–8):353–355
Salas-Orozco M, Niño-Martínez N, Martínez-Castañón GA, Méndez FT, Jasso MEC, Ruiz F (2019) Mechanisms of resistance to silver nanoparticles in endodontic bacteria: a literature review. J Nanomater 2019:7630316. https://doi.org/10.1155/2019/7630316
Samberg ME, Orndorff PE, Monteiro-Riviere NA (2011) Antibacterial efficacy of silver nanoparticles of different sizes, surface conditions and synthesis methods. Nanotoxicology 5:244–253. https://doi.org/10.3109/17435390.2010.525669
Sambrook J, Fritsch ER, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Saroj S, Shashidhar R, Bandekar J (2009) Gamma radiation used as hygienization technique for foods does not induce viable but non-culturable state (VBNC) in Salmonella enterica subsp. enterica serovar Typhimurium. Curr Microbiol 59:420–424. https://doi.org/10.1007/s00284-009-9454-3/
Shah DH, Paul NC, Sischo WC, Crespo R, Guard J (2017) Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci 96:687–702. https://doi.org/10.3382/ps/pew342
Shivaning Karabasanavar N, Benakabhat Madhavaprasad C, Agalagandi Gopalakrishna S, Hiremath J, Shivanagowda Patil G & B Barbuddhe S (2020) Prevalence of Salmonella serotypes S. Enteritidis and S. Typhimurium in poultry and poultry products. J Food Saf., 40, e12852. https://doi.org/10.1111/jfs.12852
Silver S, Le Phung T, Silver G (2006) Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J Ind Microbiol Biotechnol 33:627–634. https://doi.org/10.1007/s10295-006-0139-7
Spiliopoulou I, Zografou S, Goula A, Dimitracopoulos G, Christofidou M (2007) Molecular epidemiology and antibiotic resistance patterns of Salmonella enterica from southwestern Greece. Chemotherapy 53:392–396. https://doi.org/10.1159/000109768
Stefańska I, Rzewuska M, Binek M (2008) Evaluation of three methods for DNA fingerprinting of Corynebacterium pseudotuberculosis strains isolated from goats in Poland. Pol J Microbiol 57:105–12. https://www.researchgate.net/publication/51420221.
Stoyanova DS, Ivanova IA, Vladkova TG (2016) Nanobiotechnology against Salmonella Spp J Vet Med Res 3(4):1057. https://doi.org/10.3390/2Fmolecules21070836
Suh DK, Song JC (2006) Analysis of Salmonella enterica serotype Enteritidis isolated from human and chickens by repetitive sequence-PCR fingerprinting, antibiotic resistance and plasmid profiles. J Vet Sci 7:37–41. https://doi.org/10.4142/jvs.2006.7.1.37
Tenover FC, Arbeit RD, Goering RV (1997) How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections a review for healthcare epidemiologists. Infect Control Hosp Epidemiol 18:426–439. https://doi.org/10.1086/647644
Toker R, Kayaman-Apohan N, Kahraman M (2013) UV-curable nano-silver containing polyurethane based organic–inorganic hybrid coatings. Prog Org Coat 76:1243–1250. https://doi.org/10.1016/j.porgcoat.2013.03.023
Van Dong P, Ha CH, Binh LT, Kasbohm J (2012) Chemical synthesis and antibacterial activity of novel-shaped silver nanoparticles. Int Nano Lett 1:1–9. https://doi.org/10.1186/2228-5326-2-9
Vernet G, Mary C, Altmann DM, Doumbo O, Morpeth S, Bhutta ZA, Klugman KP (2014) Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg Infect Dis 20:434. https://doi.org/10.3201/2Feid2003.121157
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19:6823–6831. https://doi.org/10.1093/nar/19.24.6823
Washington Winner Allen SD (2001) Janda WM, koneman Paul C, Gary W & Procop GLW (2001) Color atlas and textbook of diagnostic microbiology, 6th edn. Lippincott Williams and wikinspress, Philadilphia
Wen SC, Best E, Nourse C (2017) Non-typhoidal Salmonella infections in children: review of literature and recommendations for management. J Paediatr Child Health 53:936–941. https://doi.org/10.1111/jpc.13585
WHO (2021) World Health Organization, Critically important antimicrobials for human medicine: 3rd Ed, gineva, switherland 2012. Available online: http://apps.who.int/iris/bitstream/10665/7737611/97892415044985eng.Pdf.2ua=1&ua=1. Accessed on 4 Jun 2021
WHO World Health Organization: Global antimicrobial surveillance resistance system (class) report, early implementation 2017–2018 WHO, Geneva, Switzerland 2018. Available online: http://apps.who.int/iris/bitstream/handle/10665/279636/97892415061eng.Pdf. Aaccessed on 13 Jul 2021
Wikler MA, Cockerill FR III, Bush K, Dudley MN, Eliopoulos GE, Hardy DJ, Hecht DW, Hindler JF, Patel JB, Powell M, Turnidge JD, Weinstein MP & Zimmer BL (2008) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved standard – 8th ed. Wayne, PA: Clinical and Laboratory Standards Institute, document M07-A8
Xu XH, Brownlow WJ, Kyriacou SV, Wan Q, Viola JJ (2004) Real-time probing of membrane transport in living microbial cells using single nanoparticle optics and living cell imaging. Biochemistry 43:10400–10413. https://doi.org/10.1021/bi036231a
Zahran S I, Bkheet AA & Torky HA (2020) Salmonella species isolated from different sources with special reference to biofilm formation. Alex J Vet Sci 64. https://doi.org/10.5455/ajvs.70382
Zhang Y, Peng H, Huang W, Zhou Y, Yan D (2008) Facile preparation and characterization of highly antimicrobial colloid Ag or Au nanoparticles. J Colloid Interf Sci 325:371–376. https://doi.org/10.1016/j.jcis.2008.05.063
Zhang Y, Chen Y, Zhang H, Zhang B, Liu J (2013) Potent antibacterial activity of a novel silver nanoparticle-halloysite nanotube nanocomposite powder. J Inorg Biochem 118:59–64. https://doi.org/10.1016/j.jinorgbio.2012.07.025
Acknowledgements
We wish to thank the laboratory branches, Animal Health Research Institute, Alexandria and Behera provinces, that provide facilities and Faculty of Science for electron microscopy.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
All authors have read and approved the manuscript. H. A. T., S. A. K., and R. G. T. designed the in vivo experiments. H. A. T., S. A. K., and E. K. S. reviewed the manuscript. R. G. T. wrote the manuscript. E. M. E. wrote the manuscript. E. K. S., R. G. T., A. A. B., S. K. E., H. S. A., S. I. Z., H. A. E., A. M. N., and E. M. E. performed the data analysis.
Corresponding author
Ethics declarations
Ethical approval
The current study was approved by the Ethical Committee for live birds sampling at the Faculty of Veterinary Medicine, Alexandria University, Egypt (220129).
Guidelines
All methods were carried out in accordance with relevant guidelines and regulations.
ARRIVE guidelines
The authors confirm that the study was carried out in compliance with the ARRIVE guidelines.
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Torky, H.A., Khaliel, S.AE., Sedeek, E.K. et al. Silver nanoparticle effect on Salmonella enterica isolated from Northern West Egypt food, poultry, and calves. Appl Microbiol Biotechnol 106, 5701–5713 (2022). https://doi.org/10.1007/s00253-022-12102-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12102-x