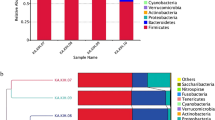
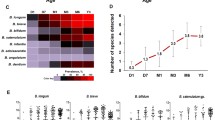
Abstract
Bifidobacteria are among the first microbial colonisers of the human infant gut post-partum. Their early appearance and dominance in the human infant gut and the reported health-promoting or probiotic status of several bifidobacterial strains has culminated in intensive research efforts that focus on their activities as part of the gut microbiota and the concomitant implications for human health. In this mini-review, we evaluate current knowledge on the genomics of this diverse bacterial genus, and on the genetic and functional adaptations that have underpinned the success of bifidobacteria in colonising the infant gut. The growing interest in functional genomics of bifidobacteria has also created interest in the interactions of bifidobacteria and their (bacterio)phages. While virulent phages of bifidobacteria have yet to be isolated, the incidence of integrated (pro)phages in bifidobacterial genomes are widely reported and this mini-review considers the role of these so-called bifidoprophages in modulating bifidobacterial populations in the human gastrointestinal tract and the implications for existing and future development of probiotic therapies.

Similar content being viewed by others
References
Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M (2012) Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7(9):e44229. https://doi.org/10.1371/journal.pone.0044229
Bottacini F, O’Connell Motherway M, Casey E, McDonnell B, Mahony J, Ventura M, van Sinderen D (2015) Discovery of a conjugative megaplasmid in Bifidobacterium breve. Appl Environ Microbiol 81(1):166–176. https://doi.org/10.1128/AEM.02871-14
Bottacini F, Morrissey R, Roberts RJ, James K, van Breen J, Egan M, Lambert J, van Limpt K, Knol J, Motherway MO, van Sinderen D (2017) Comparative genome and methylome analysis reveals restriction/modification system diversity in the gut commensal Bifidobacterium breve. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1289
Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, Rodriguez-Brito B, Salamon P, Rohwer F (2008) Viral diversity and dynamics in an infant gut. Res Microbiol 159(5):367–373. https://doi.org/10.1016/j.resmic.2008.04.006
Briner AE, Lugli GA, Milani C, Duranti S, Turroni F, Gueimonde M, Margolles A, van Sinderen D, Ventura M, Barrangou R (2015) Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS One 10(7):e0133661. https://doi.org/10.1371/journal.pone.0133661
Chaplin AV, Efimov BA, Smeianov VV, Kafarskaia LI, Pikina AP, Shkoporov AN (2015) Intraspecies genomic diversity and long-term persistence of Bifidobacterium longum. PLoS One 10(8):e0135658. https://doi.org/10.1371/journal.pone.0135658
Chassard C, Dapoigny M, Scott KP, Crouzet L, Del’homme C, Marquet P, Martin JC, Pickering G, Ardid D, Eschalier A, Dubray C, Flint HJ, Bernalier-Donadille A (2012) Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther 35(7):828–838. https://doi.org/10.1111/j.1365-2036.2012.05007.x
Dupuis ME, Moineau S (2010) Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to Listeria phages. Appl Environ Microbiol 76(5):1623–1632. https://doi.org/10.1128/AEM.02173-09
Duranti S, Gaiani F, Mancabelli L, Milani C, Grandi A, Bolchi A, Santoni A, Lugli GA, Ferrario C, Mangifesta M, Viappiani A, Bertoni S, Vivo V, Serafini F, Barbaro MR, Fugazza A, Barbara G, Gioiosa L, Palanza P, Cantoni AM, de’Angelis GL, Barocelli E, de’Angelis N, van Sinderen D, Ventura M, Turroni F (2016) Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol 92(12). https://doi.org/10.1093/femsec/fiw191
Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, Ferretti P, Gorfer V, Ferrario C, Milani C, Mangifesta M, Anzalone R, Zolfo M, Viappiani A, Pasolli E, Bariletti I, Canto R, Clementi R, Cologna M, Crifo T, Cusumano G, Fedi S, Gottardi S, Innamorati C, Mase C, Postai D, Savoi D, Soffiati M, Tateo S, Pedrotti A, Segata N, van Sinderen D, Ventura M (2017a) Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5(1):66. https://doi.org/10.1186/s40168-017-0282-6
Duranti S, Lugli GA, Mancabelli L, Turroni F, Milani C, Mangifesta M, Ferrario C, Anzalone R, Viappiani A, van Sinderen D, Ventura M (2017b) Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl Environ Microbiol 83(3):e02894–e02816. https://doi.org/10.1128/AEM.02894-16
Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D (2014) Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14(1):282. https://doi.org/10.1186/s12866-014-0282-7
Eshaghi M, Bibalan MH, Rohani M, Esghaei M, Douraghi M, Talebi M, Pourshafie MR (2017) Bifidobacterium obtained from mother’s milk and their infant stool; a comparative genotyping and antibacterial analysis. Microb Pathog 111:94–98. https://doi.org/10.1016/j.micpath.2017.08.014
Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O’Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M (2011) Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Factories 10(Suppl 1):S16. https://doi.org/10.1186/1475-2859-10-S1-S16
Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468(7320):67–71. https://doi.org/10.1038/nature09523
Hidalgo-Cantabrana C, Crawley AB, Sanchez B, Barrangou R (2017) Characterization and exploitation of CRISPR loci in Bifidobacterium longum. Front Microbiol 8:1851. https://doi.org/10.3389/fmicb.2017.01851
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962):167–170. https://doi.org/10.1126/science.1179555
Hoyles L, McCartney AL, Neve H, Gibson GR, Sanderson JD, Heller KJ, van Sinderen D (2014) Characterization of virus-like particles associated with the human faecal and caecal microbiota. Res Microbiol 165(10):803–812. https://doi.org/10.1016/j.resmic.2014.10.006
Kamo T, Akazawa H, Suda W, Saga-Kamo A, Shimizu Y, Yagi H, Liu Q, Nomura S, Naito AT, Takeda N, Harada M, Toko H, Kumagai H, Ikeda Y, Takimoto E, Suzuki JI, Honda K, Morita H, Hattori M, Komuro I (2017) Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLoS One 12(3):e0174099. https://doi.org/10.1371/journal.pone.0174099
Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR (2015) Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21(10):1228–1234. https://doi.org/10.1038/nm.3950
Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M (2014) Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80(20):6383–6394. https://doi.org/10.1128/AEM.02004-14
Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiri K, van Sinderen D, Ventura M (2017a) Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18(568):568. https://doi.org/10.1186/s12864-017-3955-4
Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiri K, van Sinderen D, Ventura M (2017b) Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18(1):568. https://doi.org/10.1186/s12864-017-3955-4
Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, Duranti S, Ward DV, Ossiprandi MC, Moineau S, van Sinderen D, Ventura M (2016) Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol 18(7):2196–2213. https://doi.org/10.1111/1462-2920.13154
Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, Oyaizu H, Tanaka R (2002) Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol 68(11):5445–5451. https://doi.org/10.1128/AEM.68.11.5445-5451.2002
Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M (2017a) The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81(4):e00036–e00017. https://doi.org/10.1128/MMBR.00036-17
Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M (2013) Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol 79(14):4304–4315. https://doi.org/10.1128/AEM.00984-13
Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M (2015a) Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81(20):7078–7087. https://doi.org/10.1128/AEM.02037-15
Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, van Sinderen D, Ventura M (2017b) Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11(12):2834–2847. https://doi.org/10.1038/ismej.2017.138
Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M (2015b) Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82(4):980–991. https://doi.org/10.1128/AEM.03500-15
Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M (2016) Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82(4):980–991. https://doi.org/10.1128/Aem.03500-15
Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D (2012) Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes 36(6):817–825. https://doi.org/10.1038/ijo.2011.153
Musilova S, Modrackova N, Doskocil I, Svejstil R, Rada V (2017) Influence of human milk oligosaccharides on adherence of bifidobacteria and clostridia to cell lines. Acta Microbiol Immunol Hung 64(4):1–8. https://doi.org/10.1556/030.64.2017.029
O’Connell Motherway M, Watson D, Bottacini F, Clark TA, Roberts RJ, Korlach J, Garault P, Chervaux C, van Hylckama Vlieg JE, Smokvina T, van Sinderen D (2014) Identification of restriction-modification systems of Bifidobacterium animalis subsp. lactis CNCM I-2494 by SMRT sequencing and associated methylome analysis. PLoS One 9(4):e94875. https://doi.org/10.1371/journal.pone.0094875
O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O’Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O’Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O’Toole PW, van Sinderen D (2011) Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108(27):11217–11222. https://doi.org/10.1073/pnas.1105380108
Obermajer T, Grabnar I, Benedik E, Tusar T, Robic Pikel T, Fidler Mis N, Bogovic Matijasic B, Rogelj I (2017) Microbes in infant gut development: placing abundance within environmental, clinical and growth parameters. Sci Rep 7(1):11230. https://doi.org/10.1038/s41598-017-10244-x
Sgorbati B, Smiley MB, Sozzi T (1983) Plasmids and phages in Bifidobacterium longum. Microbiologica 6(2):169–173
Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF (2013) Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23(1):111–120. https://doi.org/10.1101/gr.142315.112
Spinelli S, Bebeacua C, Orlov I, Tremblay D, Klaholz BP, Moineau S, Cambillau C (2014) Cryo-electron microscopy structure of lactococcal siphophage 1358 virion. J Virol 88(16):8900–8910. https://doi.org/10.1128/JVI.01040-14
Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, van Sinderen D, Ventura M (2018) Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mole Life Sci 75(1):103–118. https://doi.org/10.1007/s00018-017-2672-0
Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M (2017) Glycan utilization and cross-feeding activities by Bifidobacteria. Trends Microbiol. https://doi.org/10.1016/j.tim.2017.10.001
Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M (2016) Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10(7):1656–1668 doi:https://doi.org/10.1038/ismej.2015.236
Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, Ventura M (2014) Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett 357(1):23–33. https://doi.org/10.1111/1574-6968.12509
Turroni F, van Sinderen D, Ventura M (2009) Bifidobacteria: from ecology to genomics. Front Biosci 14:4673–4684
Ventura M, Lee JH, Canchaya C, Zink R, Leahy S, Moreno-Munoz JA, O’Connell Motherway M, Higgins D, Fitzgerald GF, O’Sullivan DJ, van Sinderen D (2005) Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl Environ Microbiol 71(12):8692–8705. https://doi.org/10.1128/AEM.71.12.8692-8705.2005
Ventura M, Sozzi T, Turroni F, Matteuzzi D, van Sinderen D (2011) The impact of bacteriophages on probiotic bacteria and gut microbiota diversity. Genes Nutr 6(3):205–207. https://doi.org/10.1007/s12263-010-0188-4
Ventura M, Turroni F, Foroni E, Duranti S, Giubellini V, Bottacini F, van Sinderen D (2010) Analyses of bifidobacterial prophage-like sequences. Antonie Van Leeuwenhoek 98(1):39–50. https://doi.org/10.1007/s10482-010-9426-4
Ventura M, Turroni F, Lima-Mendez G, Foroni E, Zomer A, Duranti S, Giubellini V, Bottacini F, Horvath P, Barrangou R, Sela DA, Mills DA, van Sinderen D (2009) Comparative analyses of prophage-like elements present in bifidobacterial genomes. Appl Environ Microbiol 75(21):6929–6936. https://doi.org/10.1128/AEM.01112-09
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI (2012) Human gut microbiome viewed across age and geography. Nature 486(7402):222–227. https://doi.org/10.1038/nature11053
Zanotti I, Turroni F, Piemontese A, Mancabelli L, Milani C, Viappiani A, Prevedini G, Sanchez B, Margolles A, Elviri L, Franco B, van Sinderen D, Ventura M (2015) Evidence for cholesterol-lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Appl Microbiol Biotechnol 99(16):6813–6829. https://doi.org/10.1007/s00253-015-6564-7
Acknowledgements
JM is in receipt of a Starting Investigator Research Grant (SIRG) (Ref. No. 15/SIRG/3430) funded by Science Foundation Ireland (SFI). DvS is a member of APC Microbiome Ireland, a research centre funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant number SFI/12/RC/2273). This work was funded by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) and MIUR to MV. JM, DVS and MV are supported by a Grand Challenges Exploration grant from the Bill and Melinda Gates Foundation (Ref. No. OPP1150567).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest.
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Mahony, J., Lugli, G.A., van Sinderen, D. et al. Impact of gut-associated bifidobacteria and their phages on health: two sides of the same coin?. Appl Microbiol Biotechnol 102, 2091–2099 (2018). https://doi.org/10.1007/s00253-018-8795-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-8795-x