Abstract
Multicopy episomal plasmids in yeast, used whenever elevated levels of foreign or homologous gene expression are necessary, are known to be less stable compared to the endogenous 2-μm plasmid they are based on, at least without selective pressure. Considering that rich medium favors growth rate and, simultaneously, is less expensive than selective medium, enhancing stability in non-selective medium is extremely desirable. In this study, we changed the architecture of a multicopy model expression plasmid, creating six isoforms (same size, same DNA content but different positions and orientations of the expression block) and studied mitotic stability, copy number, as well as reporter yEGFP3 expression between isoforms. With one isoform being significantly more stable than the others and another one exhibiting elevated plasmid copy numbers in rich medium, we show that consideration of the arrangement of the plasmid elements might be crucial for productivity employing Saccharomyces cerevisiae as a host. We strongly believe that the ideal architecture has to be assessed for each case and assembly strategy has to begin by evaluating the stability of the vector backbone before insertion of the desired gene. For the plasmid set studied, yEGFP3 reporter production depends more on mitotic stability than on elevated plasmid copy numbers in a small number of cells retaining the plasmid under non-selective conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Robustness of an industrial process employing recombinant organisms seems largely dictated by abiotic process parameters. Likewise important is the contribution of the producing organism itself. As a cell-based system, the host inevitably is subject to selection (and counterselection), disfavoring non-physiological changes of its genetics and metabolism. In order to overproduce proteins or low molecular weight metabolites, homologous or heterologous, almost always genetic information needs to be transformed into the host cell. The necessary sequences in most cases make part of a plasmid vector. The stability of such a vector in the recipient cell is of primordial importance for the robustness and productivity of the production system, instability might render the validation of an industrial process questionable or even impossible (Zhang et al. 1996; Delvigne and Goffin 2014; Gustavsson and Lee 2016).
Since a long time, the yeast Saccharomyces cerevisiae is a favorable host in industrial production, not only for food and alcoholic beverages, but also for biofuels, and, in particular, for high-value pharmaceuticals and vaccines (e.g., Ferrer-Miralles et al. 2009; Martinez et al. 2012; Nielsen 2013). Well-established genetics, high growth rates, a basic capacity for eukaryotic protein processing and secretion, and the “generally recognized as safe” (GRAS) status by the US Food and Drug Administration are just some of many beneficial traits of yeasts like S. cerevisiae. In addition, yeast has a somewhat outstanding position as it can use plasmids of various types, a feature rarely found in eukaryotic organisms (Gunge 1983; Caunt et al. 1988; Da Silva and Srikrishnan 2012). In the case of heterologous production, plasmids play an important role for easy genetic manipulation. DNA can be introduced by using YCp-type plasmids (yeast centromeric plasmids carrying a chromosomal origin of replication and additional centromeric sequences for segregational stability) or YIp-type plasmids (yeast integrative plasmids for genomic integration), both resulting in high stability. In both cases, expression levels are, however, generally low due to the presence of only one copy of the plasmid and thus of the gene of interest (Caunt et al. 1988; Romanos et al. 1992). In addition, integration might disrupt cellular functions and cause unfavorable regulatory control issues (Parker and DiBasio 1987). As an alternative, YEp-type plasmids (yeast episomal plasmids) do exist, carrying sequences of the yeast endogenous 2-μm plasmid. This 2-μm plasmid is found in most wild-type and laboratory S. cerevisiae strains (for a recent review on 2-μm structure and functions, see Chan et al. 2013). YEp-type plasmids are present in multiple copies and thus promise a benefit from a gene-dosage effect in terms of an increased productivity (Romanos et al. 1992; Da Silva and Srikrishnan 2012). The downside of using YEp-type vectors with an elevated copy number is a reduced stability. Instability can be either structural or segregational, the latter caused by uneven partitioning of plasmids during cell division (Caunt et al. 1988). Research often employs yeast as a model system in a multitude of applications. Here too, YEp-type plasmids are frequently used, though their stability seems of minor concern and is rarely questioned. Nevertheless, problems linked to selection and counterselection apply also on a small scale. Taking those problems in consideration, experimental results sometimes might ask for a second view.
Attempts have been made to increase stability and optimize productivity of YEp-type plasmids. Environmental approaches for enhancing stability include media formulation and fermentation conditions and have been discussed in detail (Zhang et al. 1996; Caunt et al. 1988). Using genetic approaches such as different yeast selectable markers and promoters, vector sets for optimized metabolic engineering have been constructed (e.g., Fang et al. 2011; Karim et al. 2013; Seresht et al. 2013; Gnügge et al. 2016; reviewed in Da Silva and Srikrishnan 2012). Complementation of chromosomal auxotrophic markers with a functional copy of the gene contained on the plasmid would allow a transformed strain to grow in selective minimal media (Sherman 1991). Likewise, a yeast-adapted gene carried on the plasmid could confer a resistance to antibiotics. Both systems are commonly used in small scale and result in high stability while maintaining a high copy number. In any case, both conditions are not feasible in large-scale production due to high costs and, in the latter case, an intensive downstream processing (Gustavsson and Lee 2016).
In continuation to a previous study, expression model vectors were constructed based on a published YEp-type multicopy plasmid (plasmid isoform construct [pIFC]3.13; Hohnholz et al. 2017). A family of plasmids varying in their architecture was assembled by addition of a yEGFP3-based reporter gene in different positions and orientations. This enabled us to study the potential impact of plasmid-derived (heterologous) gene expression on plasmid loss, plasmid copy numbers (PCNs), and levels of reporter protein expression. GFP or its variants are frequently used in cell and molecular biology studies, also in S. cerevisiae. Fluorescent proteins are thought to be without major physiological consequences for the host cell even at high expression levels. Expression of our reporter is under control of the strong constitutive yeast TEF1 promoter (Nacken et al. 1996; Sun et al. 2012; Lee et al. 2013; Peng et al. 2015), and transcription is terminated by the short synthetic Tsynth8 sequence (Curran et al. 2015) (Fig. 1a). To our knowledge, this is the first time that architectural rearrangements have been analyzed systematically to investigate stability and reporter protein expression in yeast. Unexpectedly and despite being isoforms, the arrangement of plasmid elements has an impact on mitotic stability, copy number, and protein expression in non-selective media. We demonstrate thus that it can be of significance to modify the arrangement of functional elements of an expression plasmid. Our data suggest that one isoform, pIFC4.134, represents a particularly favorable arrangement of the functional sequences, combining high stability and the highest reporter protein expression under non-selective conditions. The percentage of transformed cells in a culture seems more relevant to overall heterologous productivity than the contribution of a small number of cells carrying a high PCN.
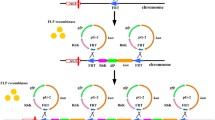
Isomeric forms of an E. coli-yeast shuttle vector (2 μm, HIS3) carrying a synthetic yEGFP3 expression block
a yEGFP3 expression block with the TEF1 promoter (TEF1
prom
), the yEGFP3 coding sequence, and the artificial transcription termination sequence Tsynth8 (Curran et al. 2015). The expression block is flanked by SacI recognition sequences.  , schematic drawing of the expression block, where the arrow marks the direction of transcription. b pIFC4.131 with the SacI sites indicating fragments borders, as well as asymmetric sites which allow to distinguish fragment orientation.
, schematic drawing of the expression block, where the arrow marks the direction of transcription. b pIFC4.131 with the SacI sites indicating fragments borders, as well as asymmetric sites which allow to distinguish fragment orientation.  , bacterial fragment (1924 bp) with oriV and bla, where the arrow marks the direction of transcription of the bla gene;
, bacterial fragment (1924 bp) with oriV and bla, where the arrow marks the direction of transcription of the bla gene;  , yeast 2 μm fragment (1405 bp), where the arrow marks the direction of transcription of the FLP gene;
, yeast 2 μm fragment (1405 bp), where the arrow marks the direction of transcription of the FLP gene;  , S. cerevisiae HIS3 gene (1017 bp), where the arrow marks the direction of transcription;
, S. cerevisiae HIS3 gene (1017 bp), where the arrow marks the direction of transcription;  , yEGFP3 expression block. c pIFC4.13X plasmid family (schematic drawing); pIFC3.13 is 4.346 bp long, pIFC4.131–4.136 plasmids are 5526 bp long. d pIFC4.112, expression vector, derived from pIFC3.11 (Hohnholz et al. 2017)
, yEGFP3 expression block. c pIFC4.13X plasmid family (schematic drawing); pIFC3.13 is 4.346 bp long, pIFC4.131–4.136 plasmids are 5526 bp long. d pIFC4.112, expression vector, derived from pIFC3.11 (Hohnholz et al. 2017)
Materials and methods
A family of isomeric multicopy model expression plasmids of 5526 bp was created based on pIFC3.13, a particularly stable member of the aforementioned pIFC3.X series (Hohnholz et al. 2017). These plasmids carry an expression block with the yeast-adapted yEGFP3 serving as reporter protein.
Escherichia coli and yeast strains
For cloning and DNA amplification, E. coli strain DH5α (Life Technologies, Darmstadt, Germany) was used employing standard molecular biology protocols. As we did not see a difference in transformation efficiency and segregational stability between the two common laboratory S. cerevisiae strains SY922 and BY4742 (Hohnholz et al. 2017), we focused in this study on the widely employed BY4742 (MAT α, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0; Brachmann et al. 1998; Euroscarf collection, Frankfurt, Germany). E. coli and yeast transformation and propagation were done as described previously (Hohnholz et al. 2017).
Assembly of the yEGFP3 expression block and plasmid construction
The yEGFP3 expression block (1180 bp; Fig. 1a) consists of a fragment of the strong constitutive yeast TEF1 promoter, a yeast-adapted synthetic sequence encoding a mutated version of the Aequorea victoria GFP (S65G, S72A; Cormack et al. 1997), and the Tsynth8 synthetic transcription termination sequence (Curran et al. 2015). TEF1 promoter sequences (YPR080W; 407 bp) were derived by PCR from yeast strain SY992 (Tomlin et al. 2001) with a SacI site added to the 5′ end. yEGFP3 sequences (714 bp) were amplified from pUG34 (supplied by J. H. Hegemann, Heinrich-Heine-Universität, Düsseldorf, Germany) adding a TAG stop codon to their 3′ end. The Tsynth8 synthetic terminator (49 bp), preceded by an A, with 6 bp added to its 3′ end for a SacI site, were assembled from a pair of oligonucleotides. Oligonucleotides used in the assembly are
RH007 5′-TATGAGCTCCCACACACCATAGCTTCAAAATG-3′,
RH008 5′-AATTCTTCACCTTTAGACATCTTAGATTAGATTGCTATGCTTTCTTTCTAATGAGC-3′,
RH009 5′-GCATAGCAATCTAATCTAAGATGTCTAAAGGTGAAGAATTATTCACTGGTG-3′, and RH010 5′-TATGAGCTCTTTGAAAGATGATACTCTTTATTCCTACATAAGTAAATGAGTTTATATATCTATTTGTACAATTCATCCATACCATGGGTAATACC-3′. The yEGFP3 expression block was inserted in pUC18 from where it was recovered as a SacI-SacI fragment.
Cloning of the expression plasmids pIFC4.131–4.136 and yeast transformation
The SacI fragment of the yEGFP3 expression block was cloned into pIFC3.13 (Hohnholz et al. 2017) partially digested with SacI. The complete series of isomeric expression plasmids was recovered with the reporter expression block inserted in all three possible positions and, in each case, in both orientations (Fig. 1c). The yEGFP3 expression block was also inserted into pIFC3.11 in the same manner, recovering pIFC4.112. For the purpose of this work, we transformed yeast strain BY4742 with each member of the series (Fig. 1c) employing a published method (Gietz and Schiestl 2007).
Plasmid loss studies
His+ transformants of each plasmid were cultivated for 16 h in selective SDsup (fully synthethic media with dextrose as carbon source and with L-leucine, L-lysine, and uracil as supplements) and for three consecutive cycles of 24 h of cultivation (= 72 h) in SD sup starting with a 1:1000 dilution or in YPDAU (YPD supplemented with adenine and uracil) starting with a 1:4000 dilution as described for previous plasmid loss studies of the pIFC3.X plasmid family unless otherwise stated (Hohnholz et al. 2017). Plasmid loss rates were calculated in the published manner (Hohnholz et al. 2017). As we expected segregational stability to drop for the expression vectors, we followed plasmid loss for a reduced time, i.e., 3 × 24 h = 72 h instead of 5 × 24 h = 120 h (in our case more than 30 generations).
Plasmid copy numbers
DNA was prepared, and PCNs were assessed by qPCR with the PerfeCTa® SYBR® Green FastMix (Quanta, Beverly, MA, USA). Detection system and analysis software were the Mastercycler® RealPlex2 and realplex software version 2.2 (Eppendorf, Hamburg, Germany). A 113 bp fragment of the HIS3 ORF (YOR202W) contained in the plasmid was amplified, likewise a 111 bp fragment of the chromosomal single copy gene ENB1 (YOL158C) serving as internal standard (Hohnholz et al. 2017).
Reporter gene expression analysis
His+ transformants (a mixture of 10 colonies randomly chosen) were recovered from a SDsup agar plate and were grown overnight in liquid SDsup at 30 °C or for 48 h in YPDAU as described for the plasmid loss studies. Cells were harvested, washed, and suspended in a 1:1 ratio of cells to potassium phosphate buffer (50 mM, pH 6.0) containing a protease inhibitor mix (Complete, Mini, EDTA-free, Roche Diagnostics, Mannheim, Germany). Glass beads (Ø 0.25–0.5 mm, Carl Roth, Karlsruhe, Germany) were added to the suspension. Cells were lysed by vortexing at Vmax in 6 cycles of 2 min with intermittent 1 min cooling on ice. Extracts were recovered and glass beads were washed twice with 50 μL of ice cold buffer. Extracts and washings were pooled and centrifuged at 13,000 x g at 4 °C for 1 min. Supernatants were withdrawn and kept at −18 °C in aliquots for further use. Protein concentrations were assessed employing the Bradford protocol with bovine serum albumin as a standard.
Native polyacrylamide gel electrophoresis (PAGE) was carried out following the protocol of Bio-Rad (Bio-Rad Laboratories GmbH, Munich, Germany). Samples with the desired amount of protein were diluted in sample buffer (Bio-Rad) and were separated on commercial 4–20% polyacrylamide gradient gels (Bio-Rad) in running buffer (Tris-glycine; following the Bio-Rad protocol). Fluorescent bands were detected illuminating the gels with the Blue/Green LED Transilluminator (Nippon Genetics, Dueren, Germany) at 480–530 nm.
Relative fluorescence was determined using the Hitachi F-2500 Fluorescence Spectrophotometer (FL Solutions program; Hitachi High-Technologies Europe GmbH, Krefeld, Germany) with an excitation of 485 nm and an emission spectrum between 500 and 560 nm. For whole cell analyses, 5 OD600 units of cells were centrifuged, washed, and resuspended in 1.5 mL water. For protein extract analyses, 80 and 40 μg of total protein were diluted in 1.5 mL of water, respectively.
Results
Growth retardation and plasmid loss
Transformation efficiencies of the plasmid isoforms did not vary significantly, and, more importantly, with 1.1 × 106 transformants per μg of plasmid DNA per 108 cells, efficiency did not decrease compared to the parental pIFC3.13 and was in the range of published results (Gietz and Schiestl 2007; Mitrikeski 2015).
Results of growth behavior of His+ transformants and plasmid loss are summarized in Table 1. Growth was retarded by at least 10% when comparing the parental vector pIFC3.13 with the expression plasmids. Plasmid loss rate only doubled in the best of all cases (pIFC4.134, Table 1), for the other isomers loss rates increased by four to fivefold with less than 25% of His+ clones after about 30 generations of growth in YPDAU. Thus, the pIFC4.13X expression plasmids cause a significant slowdown in growth, and they exhibit a marked segregational instability in our plasmid loss protocol.
As mentioned above, environmental factors affect plasmid stability. Media composition is one of those factors. In selective media, segregation stability of our isoforms is high and does not differ between the plasmids (81.5–86.7% His+ cells after 48 h in SDsup; Fig. 2a).
Copy number of model expression plasmids of the pIFC family
His+ transformants of strain BY4742 were pregrown in SDsup overnight (t0), corresponding to about 2–3 generations. Cells were then transferred to fresh SDsup or YPDAU and were then cultivated for an additional 48 h with a change of media after 24 h, corresponding to additional 7–8 generations in SDsup and 10–11 generations in YPDAU according to the plasmid loss protocol. DNA was prepared, and PCNs were determined as described in Materials and Methods. Standard deviations for the percentage of His+ clones (n = 3) were calculated as described in Table 1. PCNs shown are the mean results of three His+ clones with two biological and two technical replicates (± standard deviation). a His+ cells (in %) as determined by replica plating on selective media after preculturing in SDsup overnight and subsequent propagation for 48 h in SDsup or in YPDAU; b PCN after cultivation overnight in SDsup (t0) and subsequent propagation for 48 h in YPDAU
For expression plasmid pIFC4.112 (Fig. 1d), derived from a different backbone (pIFC3.11; Hohnholz et al. 2017), His+ clones made up less than 5% of the population after 32 generations in YPDAU.
PCNs
In rich media, PCNs generally were lower than in SDsup except for clones carrying pIFC4.133 (Fig. 2b). PCNs of the latter were more than two times higher in rich media compared to the average PCN of 2.4 (± 1.5) of the other five isoforms. PCNs did not vary to a larger extend between transformants carrying one of the pIFC4.13X vectors when the cells were propagated in selective conditions. With an average of 4.3 (± 1.0), PCNs in cells grown in SDsup were, however, more than seven times lower when compared to the parental pIFC3.13 (Fig. 2b).
We also reasoned that only His+ clones carry the respective plasmid, thus we recalculated the PCNs for the fraction of transformed cells in the population, e.g., if PCN was determined to be 10 (per haploid genome), but only 25% of cells were His+, those cells carried indeed 40 plasmids and 75% no plasmid (i.e., had lost the plasmid). This calculation led to a three-fold increase in PCN in plasmid-carrying cells, particularly pronounced in cells with the mitotically unstable plasmids pIFC4.132-33 and 35-36 (data not shown).
Reporter expression analyses
Cell-free extracts were analyzed for their relative fluorescence and thus for their content in yEGFP3 (Fig. 3a, b). The signal obtained is specific for yEGFP3 production as extracts from clones carrying the parental vector pIFC3.13 did not exhibit any detectable fluorescence. Among the six isoforms, pIFC4.134 allowed the highest productivity after 48 h in YPDAU with a 67% more intense fluorescence signal compared to pIFC4.132. When intact cells were analyzed, the results mirrored those of the extracts (data not shown). Expression levels for cells grown in SDsup did vary by less than two fold comparing the six isomers. The same is true for cells grown in YPDAU, though their expression levels were generally lower (Fig. 3a). This analysis revealed that pIFC4.131 lead to an increased productivity in selective conditions by around 59% compared to pIFC4.132 and pIFC4.133 which showed the weakest fluorescence in the protein extracts (Fig. 3a).
yEGFP3 produced from pIFC plasmids
His+ transformants of strain BY4742 were grown as described in legend to Fig. 2; extracts were prepared, and their protein content was determined as described. Extracts from yeast clones with pIFC3.13 (no expression block) served as negative control. Extracts (a) were analyzed for their relative fluorescence as described, the mean of two technical replicates (± SD) is shown. Extracts from cells grown in YPDAU were separated by native PAGE (b) as described. Extracts from yeast clones with pIFC3.13 (no expression block) served as negative control (lane 7). Gels were exposed to LED (480–530 nm) as described. Pictures were taken (B, upper panel, shown is a picture which was enhanced (Microsoft Office PowerPoint 2007) for brightness and contrast equally across the entire image) and were converted to their negative form (presented as a black and white image for increased visual sensitivity). The camera was a Pentax MX-1 (Ricoh Imaging, Hamburg, Germany) and the image processing software used (B, lower panel) was ImageJ (NIH, Bethesda, MD, USA)
Native PAGE indicated the presence of fluorescent molecules in those extracts at very low amounts of protein with pIFC4.134 giving a somewhat stronger signal than the rest (Fig. 3b, lanes 4, 11). Even at very high amounts of proteins, extracts of pIFC3.13 did not reveal any fluorescence (Fig. 3b, lane 7), confirming that fluorescence signals are linked to the presence of the expression plasmids. In PAGE, the fluorescent signal of yEGFP3 appeared to be derived from a doublet of bands, not always clearly visible (Fig. 3b). This phenomenon has been described recently where solvent molecules get “trapped” inside the yEGFP3, producing isoforms of the protein (Glukhova et al. 2017). In Coomassie-stained gels, likewise, a doublet of bands can be detected in those extracts (not shown).
Discussion
Having observed that the arrangement of the functional segments of a simple multicopy yeast-E. coli shuttle plasmid of the YEp-type can have a significant effect on the segregational stability and PCN (Hohnholz et al. 2017), we wanted to go further in investigating the behavior of a set of isomeric expression plasmids.
In a previous study, the parental plasmid pIFC3.13 (Fig. 1) used here was found to exhibit a high segregational stability in strain BY4742 (43.6% in contrast to 4.6% of His+ cells carrying the isomeric pIFC3.21 in conditions resembling industrial cultivation, i.e., more than 60 generations in non-selective conditions) (Hohnholz et al. 2017). Choosing pIFC3.13 as a mitotically stable “backbone” for assembly of a model expression vector ensured the gain of comparable results for the plasmid isoforms harboring a reporter yEGFP3 expression block (Fig. 1a) as segregational stability was expected to drop, both due to an increased size and reporter gene expression (Parker and DiBasio 1987). In contrasts to pIFC3.13, segregational stability of pIFC3.11 was particularly low in the study mentioned (14.5% His+ cells after more than 60 generations). Insertion of the same yEGFP3 expression block (with pIFC4.112 a single form got cloned, Fig. 1d) produced another plasmid isoform to the pIFC4.13X family. BY4742 His+ transformants carrying pIFC4.112 had comparable growth rates but we observed a plasmid loss rate twice as high as the most unstable pIFC4.13X isoform (3.6% His+ cells after 32 generations; Table 1), hence insertion of the expression block further aggravated the segregational instability.
All members of the set (and also pIFC4.112) caused a noticeable growth retardation in non-selective conditions in S. cerevisiae (Table 1). Likewise, an increased plasmid loss could be observed. Nevertheless, one plasmid, i.e., pIFC4.134 ( ; see legend to Fig. 1 for explanation of the arrangement of the plasmid elements) performed significantly better than the other five. Plasmid loss rate was 2.6 times lower when compared to pIFC4.135 (
; see legend to Fig. 1 for explanation of the arrangement of the plasmid elements) performed significantly better than the other five. Plasmid loss rate was 2.6 times lower when compared to pIFC4.135 ( ) and still 1.8 times lower compared to the second most stable plasmid pIFC4.136 (
) and still 1.8 times lower compared to the second most stable plasmid pIFC4.136 ( ) of the pIFC4.13X series after 30 or more generations.
) of the pIFC4.13X series after 30 or more generations.
For all six plasmids of the set, PCNs did not vary in SDsup; they dropped, however, markedly in both selective and non-selective conditions when compared to the parental pIFC3.13 (Fig. 2b). A small increase in size (of 27%) seems an unlikely explanation for the reduction as well as the observed growth retardation following the argument of Coppella and Dhurjati (1989). Therefore, the lower PCN must be the result of the addition of the yEGFP3 expression block to the plasmids. Its expression and subsequent reporter protein production seem to cause an adverse effect on the host favoring cells with a low PCN. A similar drop in copy numbers in His+ cells grown YPDAU in comparison to SDsup has been observed for the set of isomeric basic pIFC3.X plasmids (Hohnholz et al. 2017).
Using rich media in large-scale production is the only economical option. However, in research and pilot-scale fermentation processes with S. cerevisiae as a host, employing selective media might be preferred. Reduced growth rates in those (data not shown) are balanced out by a high mitotic stability that in our case showed no differences between the isoforms (Fig. 2a).
Fluorescent yEGFP3 was chosen as a quantifiable reporter (of small size) for pIFC4.13X plasmid performance. In selective conditions, pIFC4.131 allowed for the highest reporter production in the plasmid family though differences between maximum and minimum production do in general not exceed 67% in YPDAU and 59% in SDsup (Fig. 3a). Systematically, relative specific fluorescence derived from the pIFC4.13X plasmid family in cell-free extracts is two to threefold lower when transformants were propagated in YPDAU compared to those grown in SDsup (Fig. 3a). Here, highest reporter production was with pIFC4.134 (Fig. 3a, b). The decrease in reporter production from cells grown in SDsup to those grown in YPDAU seems to follow the drop in PCN with the exception of cells carrying pIFC4.133 when grown in non-selective conditions. Despite its relatively high copy number, cells with pIFC4.133 do not produce significantly more of the reporter in rich media pointing to limiting factors beyond the gene-dosage effect confirming published observations (Lee et al. 2015). pIFC4.134 seems to present a special case as fluorescence in extracts drops as for the other members of the set comparing SDsup and YPDAU grown cells whereas its PCN increases. Furthermore, this plasmid stands out for its segregational stability in non-selective conditions (Table 1).
Overall, for the pIFC4.13X plasmid family, copy number and productivity seem to be linked to a certain degree. An average, PCN masks a potential uneven distribution in a population of cells (Caunt et al. 1988; Nevoigt et al. 2006) where a small fraction of transformants with particularly high PCNs cannot reach the same productivity as a larger number of plasmid-carrying cells with a lower PCN as they risk to run into limitations from metabolic resource shortage in protein production. We reason that to some extent, productivity and PCN are proportionally connected, however the number of transformed cells in the population seems to be the determinant for productivity rather than to their PCN as can be seen in the case of pIFC4.134 (Figs. 2 and 3). High-level production of any protein might have a toxic effect on the host cell. Fluorescent proteins are widely employed in this kind of study (e.g., Soboleski et al. 2005). Nevertheless, conflicting data exist indicating deleterious effects of fluorescent protein expression causing critical levels of hydrogen peroxide production (Ansari et al. 2016; Ganini et al. 2017) or even potential protective effects (Bou-Abdallah et al. 2006).
For the performance of pIFC4.134 showing the highest stability and reporter protein production, no offhand explanations can be given. Position and orientation of the expression block seem to play a role, particularly when comparing pIFC4.134 ( ) to pIFC4.133 (
) to pIFC4.133 ( ) where the turn-around of the expression block enhances both stability and productivity. Thus, the architecture of the isoforms seems to influence the performance of the reporter gene, which in turn influences stability possibly through accumulation of the yEGFP3 or transcription-transcription/replication interferences with neighboring sequences. To what extent chromatin structure or functionality of transcriptional terminators potentially causing pervasive transcription contribute to performance of the isoforms remains speculative (Marczynski and Jaehning 1985; Murray and Cesareni 1986).
) where the turn-around of the expression block enhances both stability and productivity. Thus, the architecture of the isoforms seems to influence the performance of the reporter gene, which in turn influences stability possibly through accumulation of the yEGFP3 or transcription-transcription/replication interferences with neighboring sequences. To what extent chromatin structure or functionality of transcriptional terminators potentially causing pervasive transcription contribute to performance of the isoforms remains speculative (Marczynski and Jaehning 1985; Murray and Cesareni 1986).
Any high-level protein production will cause a metabolic burden to its respective host though this will not always become apparent in short time, small-scale experiments. It seems reasonable to assume that a change of the protein to be expressed will unpredictably influence PCNs and plasmid loss rates, e.g., in response to a potential specific toxicity of the gene product or an excessive demand of cellular resources. Thus, inevitably, assembly and optimization of any high-performance expression plasmid represent a case by case assessment.
In our opinion, a universal expression vector fulfilling all the requirements of a versatile research tool, easy to handle in terms of cloning and transformation and, at the same time, of a highly specific element for an industrial production system might not exist. Current molecular approaches consider promoters and terminators, selection markers, or host strains as adjusting screws to enhance the stability and productivity of an expression vector. The current work however demonstrates that plasmid stability, not PCN, dictates yEGFP3 productivity in yeast. The former in turn is influenced by the architecture of the expression plasmid. Having arbitrarily chosen the arrangement of elements on an expression plasmid and encountering high instability, and low copy numbers resulting in a low productivity, reconstructing the expression block or moving to an entirely different system might be more cumbersome than changing the architecture of the plasmid (including the backbone which might already be unfavorable).
In conclusion, changing the arrangement of the plasmid elements can boost stability and, as a consequence, productivity, carefully choosing both the backbone and the position and orientation of the expression block.
References
Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G, Harmon JW, Sun Z (2016) Cellular GFP toxicity and immunogenicity: potential confounders in in vivo cell tracking experiments. Stem Cell Rev 12:553–559
Bou-Abdallah F, Chasteen ND, Lesser MP (2006) Quenching of superoxide radicals by green fluorescent protein. Biochim Biophys Acta 1760:1690–1695
Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132
Caunt P, Impoolsup A, Greenfield PF (1988) Stability of recombinant plasmids in yeast. J Biotechnol 8:173–192
Chan K-M, Liu Y-T, Ma C-H, Jayaram M, Sau S (2013) The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence. Plasmid 70:2–17
Coppella SJ, Dhurjati P (1989) Effect of plasmid size and medium on growth kinetics and plasmid copy number in Saccharomyces cerevisiae. Bioprocess Eng 4:75–80
Cormack BP, Bertram G, Egerton M, Gow NAR, Falkow S, Brown AJP (1997) Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303–331
Curran KA, Morse NJ, Markham KA, Allison M, Wagman AM, Gupta A, Alper HS (2015) Short synthetic terminators for improved heterologous gene expression in yeast. ACS Synth Biol 4:824–832
Da Silva NA, Srikrishnan S (2012) Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res 12:197–214
Delvigne F, Goffin P (2014) Microbial heterogeneity affects bioprocess robustness: dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnol J 9:61–72
Fang F, Salmon K, Shen MWY, Aeling KA, Ito E, Irwin B, Tran UPC, Hatfield GW, Da Silva NA, Sandmeyer S (2011) A vector set for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast 28:123–136
Ferrer-Miralles N, Domingo-Espín J, Corchero JL, Vázquez E, Villaverde A (2009) Microbial factories for recombinant pharmaceuticals. Microbial Cell Fact 8:17
Ganini D, Leinisch F, Kumar A, Jiang J, Tokar EJ, Malone CC, Petrovich RM, Mason RP (2017) Fluorescent proteins such as eGFP lead to catalytic oxidative stress in cells. Redox Biol 12:462–468
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:31–34
Glukhova KF, Marchenkov VV, Melnik TN, Melnik BS (2017) Isoforms of green fluorescent protein differ from each other in solvent molecules ‘trapped’ inside this protein. J Biomol Struct Dyn 35:1215–1225
Gnügge R, Liphardt T, Rudolf F (2016) A shuttle vector series for precise genetic engineering of Saccharomyces cerevisiae. Yeast 33:83–98
Gunge N (1983) Yeast DNA plasmids. Annu Rev Microbiol 37:253–276
Gustavsson M, Lee SY (2016) Prospects of microbial cell factories developed through systems metabolic engineering. Microbial. Biotechnol 9:610–617
Hohnholz R, Pohlmann KJ, Achstetter T (2017) A set of isomeric episomal plasmids for systematic examination of mitotic stability in Saccharomyces cerevisiae. Yeast 34:267–275
Karim AS, Curran KA, Alper HS (2013) Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res 13:107–116
Lee ME, Aswani A, Han AS, Claire J, Tomlin CJ, Dueber JE (2013) Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res 41:10668–10678
Lee ME, DeLoache WC, Cervantes B, Dueber JE (2015) A highly-characterized yeast toolkit for modular, multi-part assembly. ACS Synth Biol 4:975–986
Marczynski GT, Jaehning JA (1985) A transcription map of a yeast centromere plasmid: unexpected transcripts and altered gene expression. Nucleic Acids Res 13:8487–8506
Martinez JL, Liu L, Petranovic D, Nielsen J (2012) Pharmaceutical protein production by yeast: towards production of human blood proteins by microbial fermentation. Curr Opin Biotech 23:965–971
Mitrikeski PT (2015) Pathways and mechanisms of yeast competence: a new frontier of yeast genetics. In: van den Berg MA, Maruthachalam K (eds) Genetic transformation systems in fungi, vol 1., fungal biology. Springer international publishing, Switzerland, pp 223–237
Murray JAH, Cesareni G (1986) Functional analysis of the yeast plasmid partition locus STB. EMBO J 5:3391–3399
Nacken V, Achstetter T, Degryse E (1996) Probing the limits of expression levels by varying promoter strength and plasmid copy number in Saccharomyces cerevisiae. Gene 175:253–260
Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U, Stephanopoulos G (2006) Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol 72:5266–5273
Nielsen J (2013) Production of biopharmaceutical proteins by yeast—advances through metabolic engineering. Bioengineered 4:207–211
Parker C, DiBasio D (1987) Effect of growth rate and expression level on plasmid stability in Saccharomyces cerevisiae. Biotechnol Bioeng 29:215–221
Peng B, Williams TC, Henry M, Nielsen LK, Vickers CE (2015) Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb Cell Factories 14:91. https://doi.org/10.1186/s12934-015-0278-5
Romanos MA, Scorer CA, Clare JJ (1992) Foreign gene expression in yeast: a review. Yeast 8:423–488
Seresht AK, Norgaard P, Palmqvist EA, Andersen AS, Olsson L (2013) Modulating heterologous protein production in yeast: the applicability of truncated auxotrophic markers. Appl Microbiol Biotechnol 97:3939–3948
Sherman F (1991) Getting started with yeast. Methods Enzymol 194:3–21
Soboleski MR, Oaks J, Halford WP (2005) Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB J 19:440–442
Sun J, Shao Z, Zhao H, Nair N, Wen F, J-H X, Zhao H (2012) Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng 109:2082–2092
Tomlin GC, Wixon JL, Bolotin-Fukuhara M, Oliver SG (2001) A new family of yeast vectors and S288C-derived strains for the systematic analysis of gene function. Yeast 18:563–575
Zhang Z, Moo-Young M, Chisti Y (1996) Plasmid stability in recombinant Saccharomyces cerevisiae. Biotechnol Adv 14:401–435
Acknowledgements
We are thankful for the skillful technical assistance of Irina Grotthaus and Birte Rossol. We would like to thank the editor for the helpful comments on the manuscript.
Funding
This work was funded by the City University of Applied Sciences Bremen.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hohnholz, R., Pohlmann, K.J. & Achstetter, T. Impact of plasmid architecture on stability and yEGFP3 reporter gene expression in a set of isomeric multicopy vectors in yeast. Appl Microbiol Biotechnol 101, 8455–8463 (2017). https://doi.org/10.1007/s00253-017-8558-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8558-0