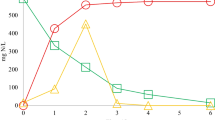
Abstract
The up-flow anaerobic sludge blanket (UASB) reactor is a promising method for the treatment of high-strength industrial wastewaters due to advantage of its high treatment capacity and settleable suspended biomass retention. Molasses wastewater as a sugar-rich waste is one of the most valuable raw material for bioenergy production due to its high organic strength and bioavailability. Interpretation for complex interactions of microbial community structures and operational parameters can help to establish stable biogas production. RNA-based approach for biogas production systems is recommended for analysis of functionally active community members which are significantly underestimated. In this study, methane production and active microbial community were characterized in an UASB reactor using molasses wastewater as feedstock. The UASB reactor achieved a stable process performance at an organic loading rate of 1.7~13.8-g chemical oxygen demand (COD,·L−1 day−1; 87–95 % COD removal efficiencies), and the maximum methane production rate was 4.01 L-CH4·at 13.8 g-COD L−1 day−1. Lactococcus and Methanosaeta were comprised up to 84 and 80 % of the active bacterial and archaeal communities, respectively. Network analysis of reactor performance and microbial community revealed that Lactococcus and Methanosaeta were network hub nodes and positively correlated each other. In addition, they were positively correlated with methane production and organic loading rate, and they shared the other microbial hub nodes as neighbors. The results indicate that the close association between Lactococcus and Methanosaeta is responsible for the stable production of methane in the UASB reactor using molasses wastewater.




Similar content being viewed by others
References
AkkoÇ N, Ghamat A, AkÇelik M (2011) Optimisation of bacteriocin production of Lactococcus lactis subsp. lactis MA23, a strain isolated from Boza. Int J Dairy Technol 64:425–432
Assenov Y, Ramírez F, Schelhorn S.-E, Lengauer T, Albrecht M 2008. Computing topological parameters of biological networks. Bioinformatics, 24, 282-284.
Azizan KA, Baharum SN, Mohd Noor N (2012) Metabolic profiling of Lactococcus lactis under different culture conditions. Molecules 17:8022–8036
Boopathy R, Tilche A (1991) Anaerobic digestion of high strength molasses wastewater using hybrid anaerobic baffled reactor. Water Res 25:785–790
Bӧrjesson P, Mattiasson B (2008) Biogas as a resource-efficient vehicle fuel. Trends Biotechnol 26:7–13
Carr FJ, Chill D, Maida N (2002) The lactic acid bacteria: a literature survey. Crit Rev Microbiol 28:281–370
Demirel B, Scherer P (2008) The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev Environ Sci Biotechnol 7:173–190
Díaz EE, Stams AJM, Amils R, Sanz JL (2006) Phenotypic properties and microbial diversity of methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl Environ Microbiol 72:4942–4949
Duncan SH, Louis P, Flint HJ (2004) Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817
Fall PA, Leroi, F., Chevalier, F., Guérin, C., Pilet, M.-F., 2010. Protective effect of a non-bacteriocinogenic Lactococcus piscium CNCM I-4031 strain against Listeria monocytogenes in sterilized tropical cooked peeled shrimp. J Aquat Food Prod Technol, 19, 84-92.
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Micro 10:538–550
Jaenicke S, Ander C, Bekel T, Bisdorf R, Droge M., Gartemann K.-H, Junemann S, Kaiser, O, Krause L, Tille F, Zakrzewski M, Puhler A, Schluter A, Goesmann A, 2011. Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS One, 6, e14519.
Janssen PH, Evers S, Rainey FA, Weiss N, Ludwig W, Harfoot CG, Schink B (1995) Lactosphaera gen. nov., a new genus of lactic acid bacteria, and transfer of Ruminococcus pasteurii Schink 1984 to Lactosphaera pasteurii comb. nov. Int J Syst Bacteriol 45:565–571
Kim TG, Moon K-E, Yun J, Cho K-S (2013) Comparison of RNA- and DNA-based bacterial communities in a lab-scale methane-degrading biocover. Appl Microbiol Biotechnol 97:3171–3181
Kim TG, Jeong S-Y, Cho K-S (2014) Functional rigidity of a methane biofilter during the temporal microbial succession. Appl Microbiol Biotechnol 98:3275–3286
Leclerc M, Delgènes J-P, Godon J-J (2004) Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ Microbiol 6:809–819
Lee J-Y, Yun J, Kim TG, Wee D, Cho K-S (2014) Two-stage biogas production by co-digesting molasses wastewater and sewage sludge. Bioprocess Biosyst Eng 1-13
Lim SJ, Kim T-H (2014) Applicability and trends of anaerobic granular sludge treatment processes. Biomass Bioenerg 60:189–202
Lin P-Y, Whang L-M, Wu Y-R, Ren W-J, Hsiao C-J, Li S-L, Chang J-S (2007) Biological hydrogen production of the genus Clostridium: metabolic study and mathematical model simulation. Int J Hydrog Energy 32:1728–1735
Noike T, Takabatake H, Mizuno O, Ohba M (2002) Inhibition of hydrogen fermentation of organic wastes by lactic acid bacteria. Int J Hydrog Energy 27:1367–1371
Ohnishi A, Hasegawa Y, Abe S, Bando Y, Fujimoto N, Suzuki M (2012) Hydrogen fermentation using lactate as the sole carbon source: solution for ‘blind spots’ in biofuel production. RSC Advances 2:8332–8340
Onodera T, Sase S, Choeisai P, Yoochatchaval W, Sumino H, Yamaguchi T, Ebie Y, Xu K, Tomioka N, Syutsubo K (2011) High-rate treatment of molasses wastewater by combination of an acidification reactor and a USSB reactor. J Environ Sci Health, Part A 46:1721–1731
Onodera T, Sase S, Choeisai P, Yoochatchaval W, Sumino H, Yamaguchi T, Ebie Y, Xu K, Tomioka N, Mizuochi M, Syutsubo K (2012) Evaluation of process performance and sludge properties of an up-flow staged sludge blanket (USSB) reactor for treatment of molasses wastewater. Int J Environ Res 6:1015–1024
Oude Elferink SJWH, Krooneman J, Gottschal JC, Spoelstra SF, Faber F, Driehuis F (2001) Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl Environ Microbiol 67:125–132
Reysenbach A-L, Longnecker K, Kirshtein J (2000) Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-Atlantic ridge hydrothermal vent. Appl Environ Microbiol 66:3798–3806
Rotaru A-E, Shrestha PM, Liu F, Shrestha M, Shrestha D, Embree M, Zengler K, Wardman C, Nevin KP, Lovley DR (2014) A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci 7:408–415
Sánchez Riera F, Córdoba P, Siñeriz F (1985) Use of the UASB reactor for the anaerobic treatment of stillage from sugar cane molasses. Biotechnol Bioeng 27:1710–1716
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schmidt JE, Ahring BK (1996) Granular sludge formation in upflow anaerobic sludge blanket (UASB) reactors. Biotechnol Bioeng 49:229–246
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Sørensen KB, Teske A (2006) Stratified communities of active archaea in deep marine subsurface sediments. Appl Environ Microbiol 72:4596–4603
Stams AJM, Plugge CM (2009) Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Micro 7:568–577
Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow C-ET, Sachdeva R, Jones AC, Schwalbach MS, Rose JM, Hewson I, Patel A, Sun F, Caron DA, Fuhrman JA (2011) Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414–1425
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen, S.J., Karlsson, A., 2013. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol, 85, 612-626.
ten Brummeler E, Pol LWH, Dolfing J, Lettinga G, Zehnder AJB (1985) Methanogenesis in an upflow anaerobic sludge blanket reactor at pH 6 on an acetate-propionate mixture. Appl Environ Microbiol 49:1472–1477
Xia L, Steele J, Cram J, Cardon Z, Simmons S, Vallino J, Fuhrman J, Sun F (2011) Extended local similarity analysis (eLSA) of microbial community and other time series data with replicates. BMC Syst Biol 5:S15
Acknowledgment
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (NRF-2012R1A2A2A03046724).
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, T.G., Yun, J. & Cho, KS. The close relation between Lactococcus and Methanosaeta is a keystone for stable methane production from molasses wastewater in a UASB reactor. Appl Microbiol Biotechnol 99, 8271–8283 (2015). https://doi.org/10.1007/s00253-015-6725-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6725-8